Top 10 discoveries about ancient people from DNA in 2022
Research on ancient genomes has moved way beyond population mixture into broader questions about how ancient people lived and interacted with their environments.

Ancient DNA was in the news this year as never before. In October, ancient DNA pioneer Svante Pääbo received the Nobel Prize in Physiology or Medicine, drawing worldwide attention on a very young scientific field. It is only 12 years since the sequencing of the first draft Neandertal and Denisovan genomes. Since then there have been dozens of major breakthroughs in our understanding of past people through their genomes.
Now ancient DNA has become a practical tool for many anthropological research topics. To be sure, researchers are still learning about extinct groups of people by sequencing genomes. But more and more new teams of scientists are using DNA to understand the context of sites and the foods that ancient people ate. And a growing frontier of research is understanding how ancient genes may contribute to traits in living people.
I've selected ten stories about ancient DNA this year that highlight this fast changing science. Some of these stories got global headlines. Others may have passed under the media radar, but I see how they are charting new avenues of information that researchers will be following over the next decade. Together they illustrate our growing knowledge of the human past.
Neandertal family ties
The Neandertals who used Chagyrskaya Cave, Russia, didn't leave much physical evidence for archaeologists. The cave has yielded a few dozen teeth and bone fragments, mostly from a layer that formed between 59,000 and 51,000 years ago. In October, a team of scientists led by Laurits Skov reported genetic data from 16 of these fragments.

What they found was detailed evidence of kinship among these bits of bone. A few individuals were represented by two or three teeth or other fragments. A vertebral fragment and a molar tooth came from a father and his daughter. The father had two other male relatives in the site, all sharing an mtDNA lineage and a unique situation called mitochondrial heteroplasmy—meaning that all three of these individuals inherited variation in the mtDNA variation within their cells from their recent maternal relatives. A finger bone and a canine tooth came from second-degree relatives, possibly an uncle and niece.
This isn't the first evidence of kinship within Neandertal sites. The El Sidrón, Spain, Neandertals have a pattern of mtDNA sharing that suggested maternal relationships among the male individuals. La Ferrassie, France, has burials in a spatial arrangement suggesting that the individuals were members of a single group.
What is new about the Chagyrskaya evidence is the ability to retrieve information of a group's members from tiny fragments that might otherwise have been missed. And it leaves a mystery: How do we have so much evidence for members of a single small group or population, with so little skeletal evidence from each of the individuals?
DNA locked in tiny poopules
More and more evidence of DNA from archaeological sites is coming from sediment samples instead of from bones or teeth. But how does this DNA survive?
In the first issue of PNAS of the year, Diyendo Massilani and collaborators transformed our understanding of where ancient DNA really comes from. They started from small sections of sediment from Denisova Cave, Russia, which they fixed in resin. They then sliced the section and sampled for DNA across its surface.
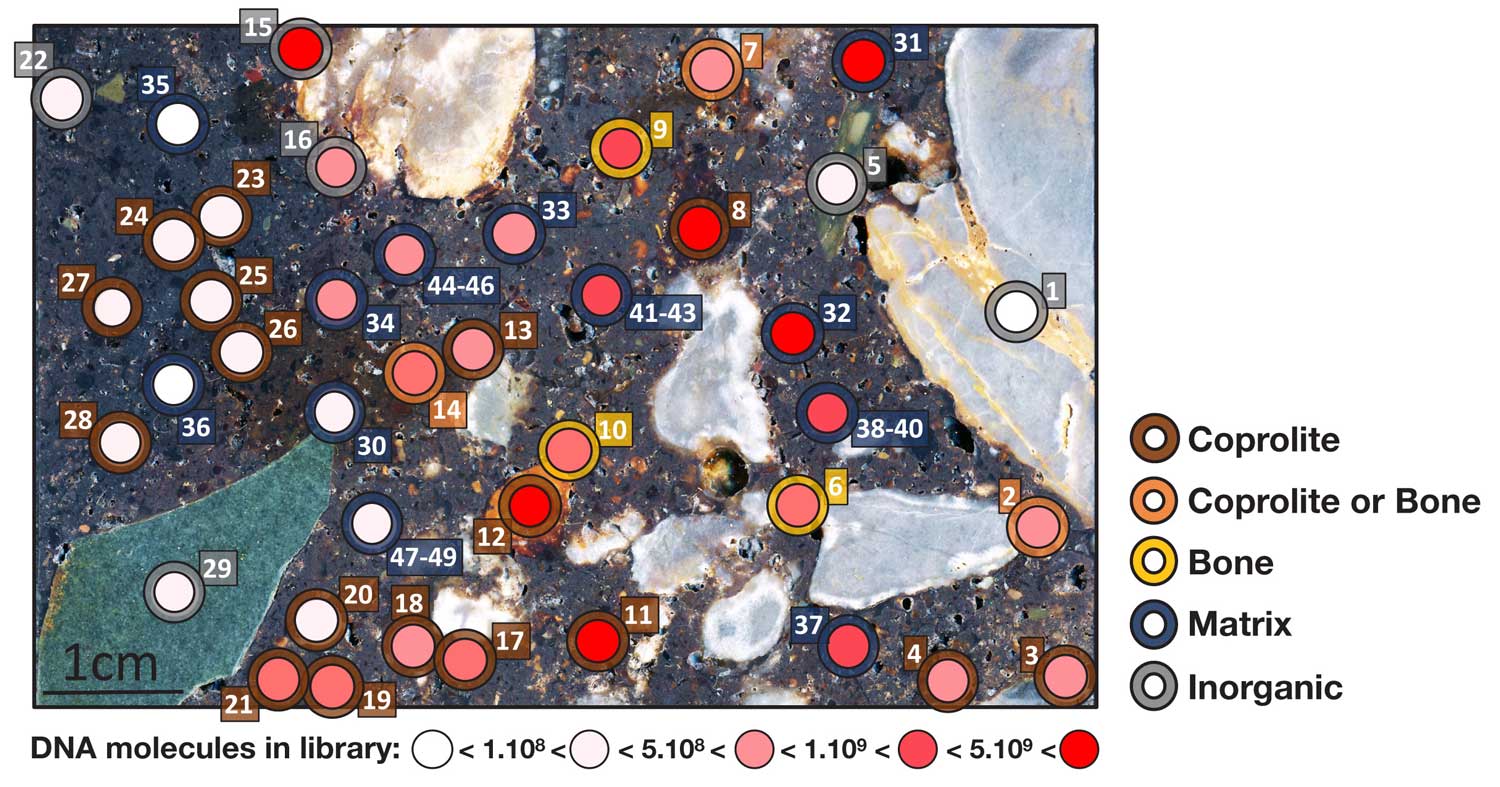
They discovered that DNA is often preserved within tiny fragments of bone or tiny fragments of coprolites—ancient fecal remains—that are scattered throughout the sediment. Its preservation can be very different in different areas of the sediment, which may reflect chemical or other variation within the site.
The study points to a new way of studying the formation of archaeological sites and the evidence they contain. It is unrealistic to sample every particle, but it's clear that some sites contain vast amounts of biological information that would once have been discarded without anyone ever looking. It will be a continuing challenge to design research that recovers more and more of this data.
Bones of ancient Greenland
Some of the biggest news from ancient DNA this year involved other species and ancient ecosystems. In December, Kurt Kjær and collaborators made a splash when they reported the oldest DNA recovery yet, more than 2 million years old, from northern Greenland. That was long before humans arrived on this island but gives intriguing details about an ecosystem from the early Pleistocene. It may give hints about the potential for surprising discoveries in archaeological contexts.
There was another paper about ancient Greenland this year that took a much more human perspective by examining ancient hunting and fishing practices by three cultures that inhabited Greenland during the last 5000 years. Frederik Seersholm and coworkers did metagenomic analysis of bulk bone samples from archaeological sites in several areas of Greenland. The bone came from sites associated with three different cultural groups: the Saqqaq, who were the earliest inhabitants, the Thule, who are immediate ancestors of the current Indigenous people, and the Norse who settled on the island in medieval times. Seersholm and his team wanted to understand how these groups may have used the island's resources differently.
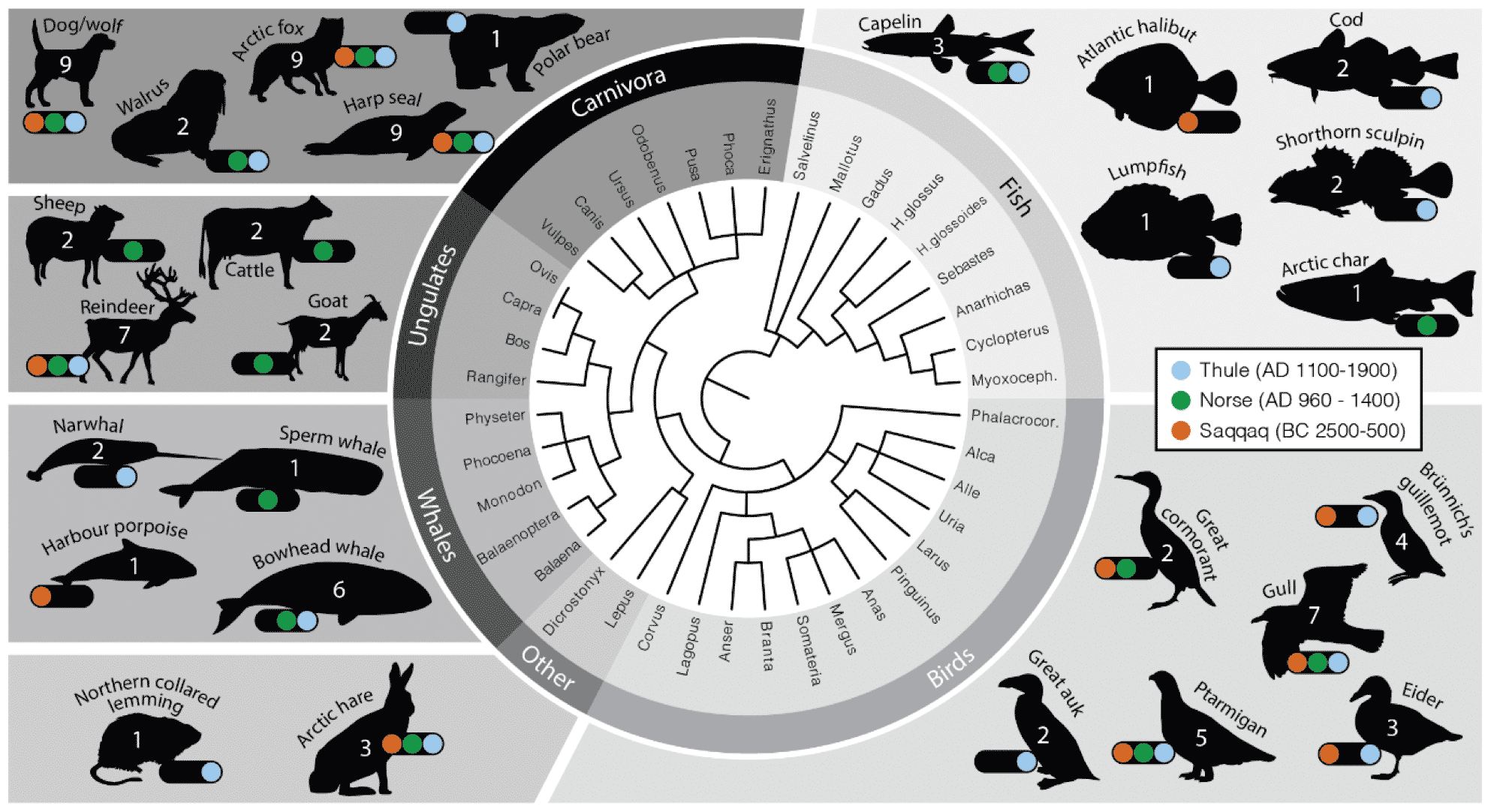
“Bulk bone samples” in an archaeological context are generally unidentifiable fragments that may come from foods or kitchen trash. This study is one of a growing literature in which ancient DNA reveals species that have usually remained invisible to archaeologists. That includes the capelin, which were a big component of both Thule and Norse diets but harvested at a small size that leaves little recognizable bone. Bowhead whales were also present only as bone fragments without identifiable anatomy, and Seersholm and coworkers were able to show their presence in sites beyond the present bowhead whale range.
The study also identified an extinct lineage of caribou in one of the ancient Saqqaq sites. This adds to the record of human impact on the faunal population after first human arrival on the island.
Probing the plant remains from ancient caves
Ancient people used many kinds of plants for food, clothing, bedding, tools, as well as for aesthetic purposes like cosmetics and decoration. But archaeologists' sources of evidence about plants from ancient sites are extremely limited. One kind of evidence is ancient pollen, but flowers and windblown pollen don't say much about the parts of plants that people often used. Some of the most valuable evidence has come from charred bits of plant material that remain in ancient fires.
Ancient DNA is a new source of evidence about ancient plants. Researchers who are examining sediment DNA can include primers for chloroplast DNA to develop evidence about plant remains in archaeological layers. In November, the Journal of Human Evolution published an article by Anneke ter Schure and coworkers, who looked at plant DNA from the layers of Aghitu-3 Cave, Armenia. They sampled layers with Upper Paleolithic artifact assemblages, between 43,000 and 26,000 years old.
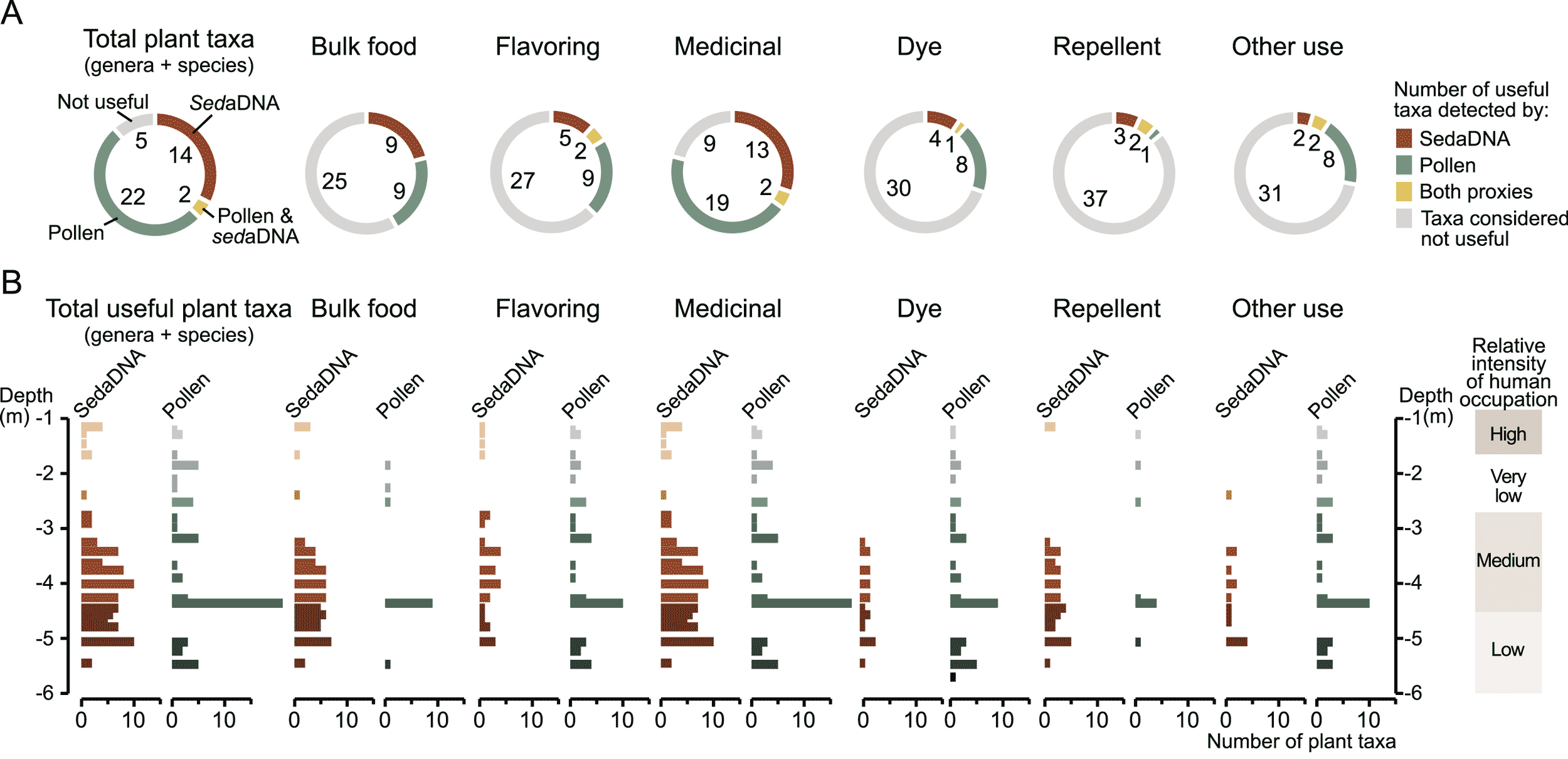
The study combined analysis of chloroplast DNA with assessments of pollen in the same archaeological layers of the site. What ter Schure and collaborators found is that sediment DNA detects many plant species and genera that are absent from the pollen record. The pollen record misses half of plant taxa that are potential foods, and nearly half of the potential medicinal plants that are present in the archaeological layers.
The pollen record is very rich in some layers and weak in others, while the sediment DNA record of plant diversity has greater evenness across layers of this site. The study shows that different kinds of evidence have different strengths. Putting them together provides a level of information that no single source of data can match.
Mixing along the African rift
The biggest gaps in our current knowledge of ancient genomes are in Africa. DNA survives better over time in cold places, and such evidence from African archaeological sites has been limited to the last 20,000 years at most. This leaves out the vast span of time when today's human populations were beginning to diversify—as well as more ancient groups such as Homo naledi.

Still, researchers have been working hard to build larger datasets from the last 20,000 years. In February, Mark Lipson and coworkers reported DNA recovery from the remains of six humans from archaeological sites in Malawi, Tanzania, and Zambia. The individuals range in radiocarbon age from more than 16,000 years ago up to the last 5000 years. They added these new samples together with some previously-sampled individuals from other sites, enabling the team to look at variation stretching from Kenya to the southern end of the Malawi rift.
Earlier work from Lipson and coworkers showed that present-day African populations descend from four early groups. In the new research, they show that three of these groups contribute ancestry to eastern African ancient people—one similar to the mid-Holocene “Mota” genome from Ethiopia, one most similar to Mbuti people from central Africa, and one most similar to southern African hunter-gatherers—in mixtures that vary from north to south.
The present gradient of variation from north to south across this region formed before 20,000 years ago. Before this there was long-distance movement connecting people across the entire region, while after this time most interactions were more local in scale and population structure became more intense.
Ancestral connections in the Bay Area
Human geneticists have given very unequal attention to different populations around the world. Most research has been done on majority populations of the U.S., western Europe, China, and Japan. Minority populations within those countries, and Indigenous people from elsewhere in the world, are underrepresented in research.
Today geneticists who aim to do good scientific research on the history of Indigenous groups engage with those communities to build relationships and give them a voice in the research questions and design. One of the strongest recent papers was published in March by Alissa Severson and collaborators. The work began with archaeological excavation of two village sites near Sunol, California, east of the San Francisco Bay. These excavations were necessary due to development, and both the excavation and later genomic work were done with participation by the Muwekma Ohlone tribe. Members attended the Summer Internship for Indigenous Peoples in Genomes program and consulted with the scientist collaborators to develop safeguards for data from research participants and for the ancient data.
The genetic results of the study showed a pattern of similarities between the current Muwekma Ohlone research participants and the remains of people found during the excavations, all from the last 2000 years. These similarities were manifested both in model-based cluster analysis and in segments of identity-by-state shared by ancient and living people.
The study also considered broader questions of relationships with other ancient sites, to examine some hypotheses of linguistic group origins and movements. The data don't answer all these questions but do show genetic continuities within and across California over time.
The study is a strong example of the kind of work that anthropological geneticists are carrying out in collaboration with Indigenous groups. Bringing communities into the research conception and design is leading to results that engage people in their history and that build a foundation for future work.
Opening a door to ancient phenotypes
Over the last decade, geneticists have probed larger and larger samples of genotypes from living people to try to understand how genes are related to traits. The most common statistical approach to relate many genotypes to phenotypes is calculation of a value for each individual known as the polygenic risk score (PRS).
This year, a group of researchers led by Samantha Cox took a big step in relating ancient DNA and ancient phenotypes. They took a look through the growing database of ancient DNA from Europe and cross-checked it against anthropological datasets of skeletal measurements. They were looking for individuals with known evidence of stature, mostly by measured femur length but sometimes based on other measurements.
They found 132 ancient individuals with both genome data and femur length estimates, and applied a PRS based on the UK Biobank study of stature to the ancient genomes. They found that the polygenic scores of the ancient individuals correlate with their actual femur lengths—suggesting that some of the genotypes that are associated with stature in today's Europeans were also responsible for variation in the past.
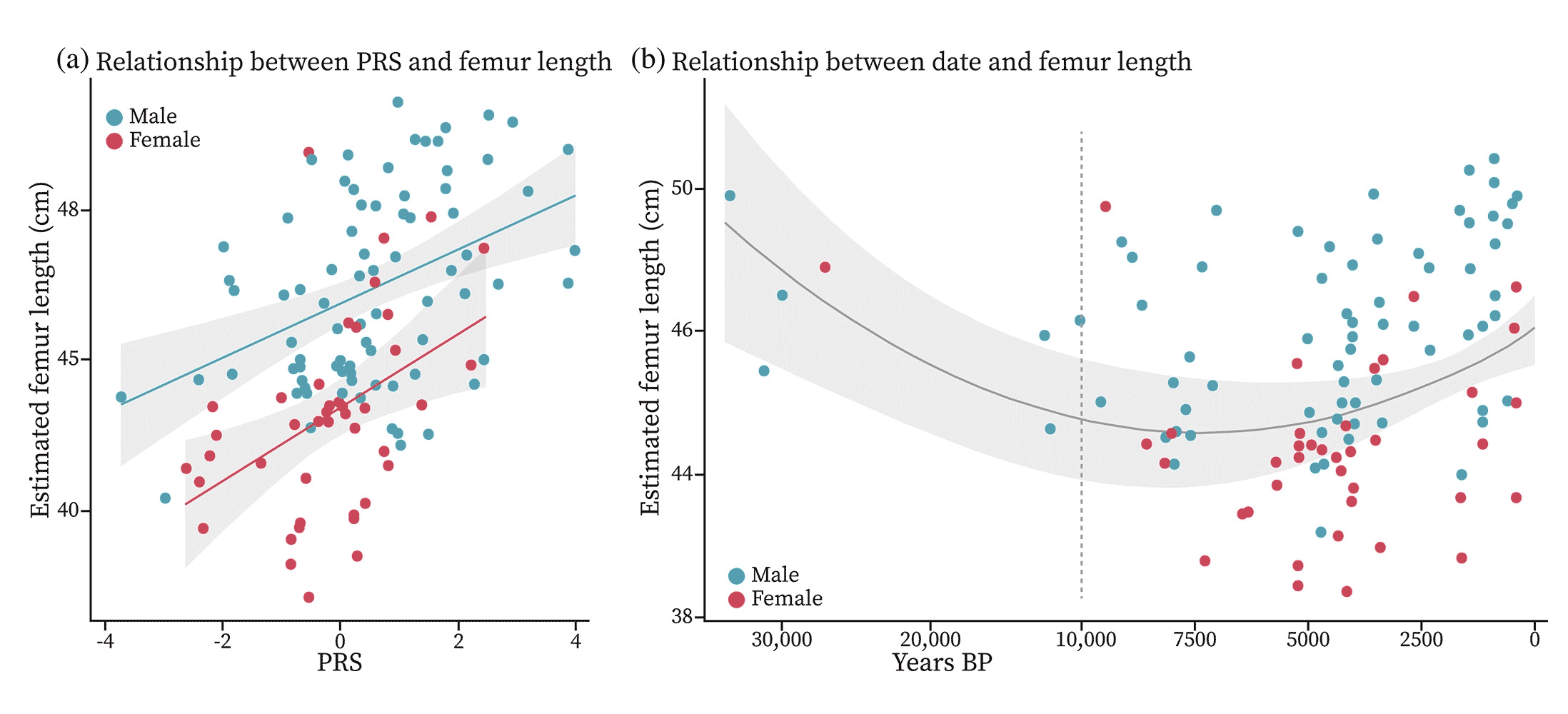
There is plenty of reason for caution in these results. We know more about the genetics of stature than almost any other continuous trait. Stature has high heritability within populations, and polygenic scores for stature include hundreds of genetic associations. But even so, the PRS estimation of femur length only explains 6.3% of variation in actual femur length for the ancient samples. That's worse than fuzzy.
Still, the PRS is almost as bad for living people as it is for the ancient ones. It's simply not a precise way of estimating the stature of any particular ancient skeleton. What it can do is provide some insight into how the genes that affect stature today have evolved over time. As datasets improve for other skeletal features across living human groups, ancient DNA will become more useful to examine the evolution of phenotypes.
Footprints of ancient plagues
Many of the fastest-evolving parts of human genomes are related to immunity. That fast evolution and the maintenance of high diversity for some immune-related genes are both a result of deaths from pathogens. But most epidemics are short-term events when measured in generations, making it very difficult to study their effects on gene frequencies.
Now ancient DNA is starting to enable geneticists to look directly at the mortality and survival of people with different genotypes during ancient pandemics. None was more massive than the Black Death, which killed up to half of Europe's population between 1346 and 1352 CE.

Jennifer Klunk and coworkers looked at genotype data from individuals who were buried in the East Smithfield mass grave in central London, who died during the Black Death, compared with cemetery burials from slightly earlier and later in time. The researchers also looked at individuals in Denmark from before and after the pandemic. These samples are a time series enabling the researchers to look at two kinds of outcomes—plague victims and plague survivors—compared with individuals who had died before the Black Death.
They found several genes that exhibited strong frequency changes during and after the Black Death—genes that either helped people survive, or led to a higher risk of death. What is powerful about this study is that the researchers took one extra step: They tested immune cell lines carrying these genotypes for in vitro response to Yersinia pestis, the pathogen that caused the pandemic. These experiments showed that these genes that mattered during the Black Death caused a response in immune cells when exposed to this pathogen.
The study is a great example of how ancient DNA can demonstrate selection in conjunction with experiments to examine the function of the selected genes.
Denisovan inputs to immunity
We've now known for 12 years that Neandertals and Denisovans both contributed genes to living human populations. It has been slow for scientists to build an understanding of what difference those genes make to human biology today. One reason is that we have so much to learn about how human genes work in general—hundreds of genes may contribute to the variation in any particular trait. But when it comes to Denisovan genes, a more pressing reason is that people from island southeast Asia and Oceania who have the highest Denisovan ancestry are underrepresented in genetic research.
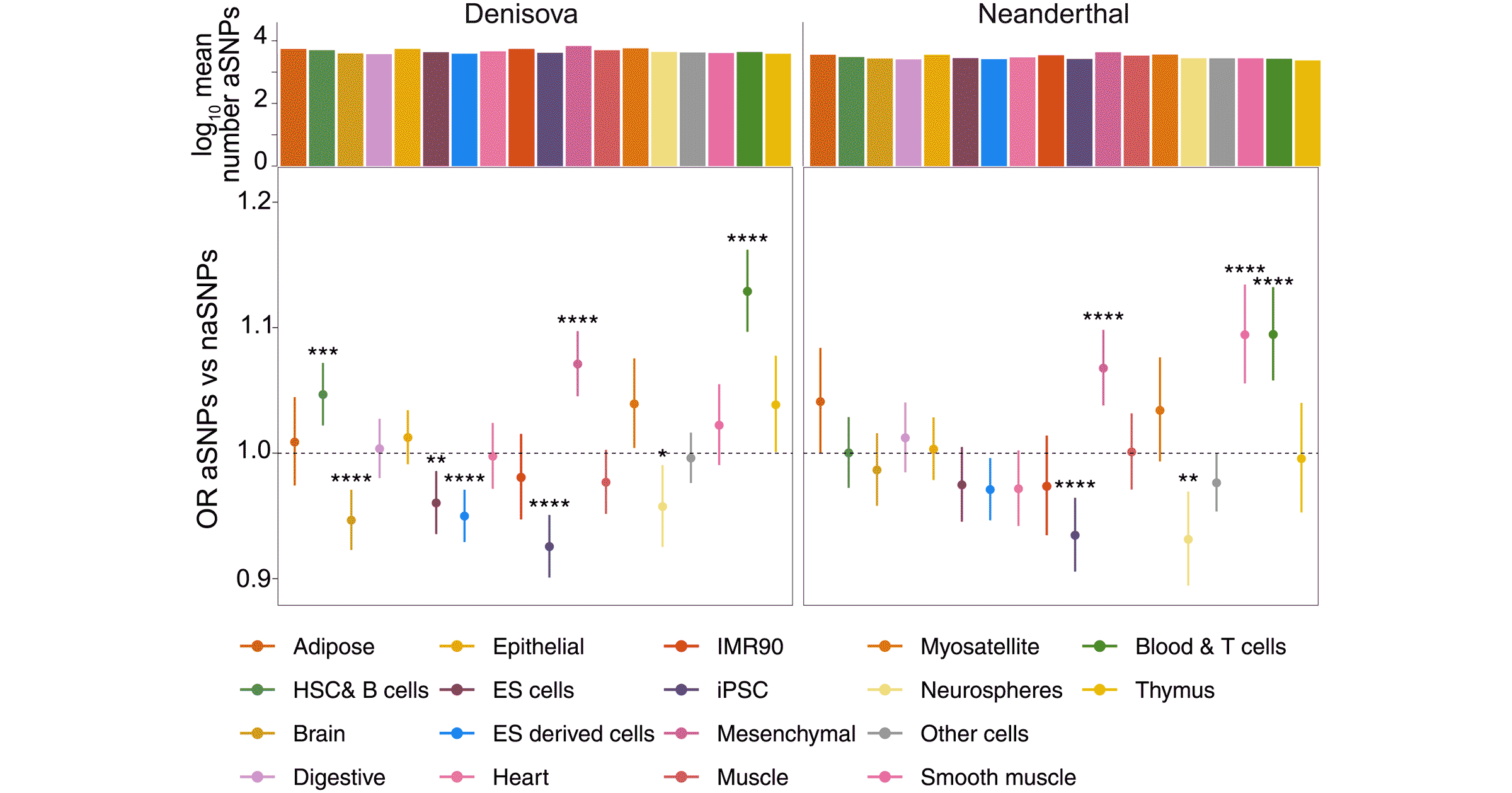
Davide Vespasiani and coworkers looked at Denisovan genes in Indonesia broadly and within Papuan people more specifically. They started from the observation that archaic genes from both Neandertals and Denisovans are less likely to impact coding sequences than non-coding sequences. From there, they built some data on the relative under- or overrepresentation of archaic alleles in various cell types. Denisovan genes in particular are likely to be active in immune cells.
They then examined some of the top candidates for adaptive introgression, particularly the OAS2 and OAS3 genes, both important to innate immune responses to viruses. They showed for the first time that the alleles inherited from Denisovans are directly involved in regulating expression of these genes.
Population flows through Anatolia
Sequencing genomes from ancient modern humans was long considered one of the greatest challenges in ancient DNA research. Neandertals were easier in many ways because they exhibit more differences from lab workers, archaeologists, and other people who might be sources of contamination. It has been under a decade since researchers reliably generated whole-genome data from Upper Paleolithic and later populations. A single ancient genome was a breakthrough in 2014.
This summer, Iosif Lazaridis and coworkers published three papers that show how far the field has advanced in sheer data generation. The combined release of these three papers includes genome data from the skeletal remains of more than 700 ancient people of Anatolia and adjacent regions.

The papers filled a hole in earlier research on DNA from more northerly archaeological sites across the steppes of eastern Europe. Over the last seven years, ancient DNA has grown to support the idea that today's widespread Indo-European language family emerged from an early Bronze Age steppe group that archaeologists know as the Yamnaya culture. But the DNA data did not yet cover Anatolia, where some of the earliest-known Indo-European languages existed.
The new work confirms a modest steppe contribution to Bronze Age Anatolian populations, and the presence of steppe-derived Y chromosome lineages in this region and in Mycenean Greece. The research also sheds light on early farming populations in Anatolia and nearby Mesopotamia, showing a complex series of interactions among Pre-Pottery and Pottery Neolithic populations of the region.
Looking into the future
This final study shows how geneticists have been filling in the global map by adding archaeological samples to growing DNA datasets. Some kinds of questions, especially concerning migration and dispersal, benefit from larger sample sizes. Can this model of large-scale DNA recovery of hundreds, or even thousands, of genomes continue into the future?
Archaeologists have spent more than a hundred years developing an understanding about how the limits of their record can affect interpretations of the past. Ancient DNA is starting to face those problems, as research transitions from questions that can be answered with a few genomes to those where sampling biases make more and more difference to the outcome.
I expect we will continue to see studies with denser sampling of times and places where skeletal remains are abundant. I also predict that some teams will redirect toward research questions that require whole-genome sequencing instead of the targeted capture of genotypes already known to vary across samples. A key to whether high investment in ancient DNA research can be sustainable is the extent to which it can address questions beyond migration and dispersal—such as the kinship record of particular sites, the utilization of resources, and the interactions of people and their environments.
This year's research shows how such a science may unfold. The methods of ancient DNA analysis are becoming essential to answer many kinds of questions.
Notes: Images from research papers here are used under terms of their Creative Commons licenses. Massilani et al. 2022 CC-BY 2.0 license from the published PNAS article. Seersholm et al. 2022 from the bioRxiv preprint. Cox et al. 2022 CC-BY-NC 4.0 license from AJPA article. Vespasiani et al. 2022 from PLoS Genetics under CC-BY 4.0 license.
References
Cox, S. L., Moots, H. M., Stock, J. T., Shbat, A., Bitarello, B. D., Nicklisch, N., Alt, K. W., Haak, W., Rosenstock, E., Ruff, C. B., & Mathieson, I. (2022). Predicting skeletal stature using ancient DNA. American Journal of Biological Anthropology, 177(1), 162–174. https://doi.org/10.1002/ajpa.24426
Kjær, K. H., Winther Pedersen, M., De Sanctis, B., De Cahsan, B., Korneliussen, T. S., Michelsen, C. S., Sand, K. K., Jelavić, S., Ruter, A. H., Schmidt, A. M. A., Kjeldsen, K. K., Tesakov, A. S., Snowball, I., Gosse, J. C., Alsos, I. G., Wang, Y., Dockter, C., Rasmussen, M., Jørgensen, M. E., … Willerslev, E. (2022). A 2-million-year-old ecosystem in Greenland uncovered by environmental DNA. Nature, 612(7939), Article 7939. https://doi.org/10.1038/s41586-022-05453-y
Klunk, J., Vilgalys, T. P., Demeure, C. E., Cheng, X., Shiratori, M., Madej, J., Beau, R., Elli, D., Patino, M. I., Redfern, R., DeWitte, S. N., Gamble, J. A., Boldsen, J. L., Carmichael, A., Varlik, N., Eaton, K., Grenier, J.-C., Golding, G. B., Devault, A., … Barreiro, L. B. (2022). Evolution of immune genes is associated with the Black Death. Nature, 611(7935), Article 7935. https://doi.org/10.1038/s41586-022-05349-x
Lazaridis, I., Alpaslan-Roodenberg, S., Acar, A., Açıkkol, A., Agelarakis, A., Aghikyan, L., Akyüz, U., Andreeva, D., Andrijašević, G., Antonović, D., Armit, I., Atmaca, A., Avetisyan, P., Aytek, A. İ., Bacvarov, K., Badalyan, R., Bakardzhiev, S., Balen, J., Bejko, L., … Reich, D. (2022). The genetic history of the Southern Arc: A bridge between West Asia and Europe. Science, 377(6609), eabm4247. https://doi.org/10.1126/science.abm4247
Lazaridis, I., Alpaslan-Roodenberg, S., Acar, A., Açıkkol, A., Agelarakis, A., Aghikyan, L., Akyüz, U., Andreeva, D., Andrijašević, G., Antonović, D., Armit, I., Atmaca, A., Avetisyan, P., Aytek, A. İ., Bacvarov, K., Badalyan, R., Bakardzhiev, S., Balen, J., Bejko, L., … Reich, D. (2022). Ancient DNA from Mesopotamia suggests distinct Pre-Pottery and Pottery Neolithic migrations into Anatolia. Science, 377(6609), 982–987. https://doi.org/10.1126/science.abq0762
Lipson, M., Sawchuk, E. A., Thompson, J. C., Oppenheimer, J., Tryon, C. A., Ranhorn, K. L., de Luna, K. M., Sirak, K. A., Olalde, I., Ambrose, S. H., Arthur, J. W., Arthur, K. J. W., Ayodo, G., Bertacchi, A., Cerezo-Román, J. I., Culleton, B. J., Curtis, M. C., Davis, J., Gidna, A. O., … Prendergast, M. E. (2022). Ancient DNA and deep population structure in sub-Saharan African foragers. Nature, 603(7900), Article 7900. https://doi.org/10.1038/s41586-022-04430-9
Massilani, D., Morley, M. W., Mentzer, S. M., Aldeias, V., Vernot, B., Miller, C., Stahlschmidt, M., Kozlikin, M. B., Shunkov, M. V., Derevianko, A. P., Conard, N. J., Wurz, S., Henshilwood, C. S., Vasquez, J., Essel, E., Nagel, S., Richter, J., Nickel, B., Roberts, R. G., … Meyer, M. (2022). Microstratigraphic preservation of ancient faunal and hominin DNA in Pleistocene cave sediments. Proceedings of the National Academy of Sciences, 119(1), e2113666118. https://doi.org/10.1073/pnas.2113666118
Seersholm, F. V., Harmsen, H., Gotfredsen, A. B., Madsen, C. K., Jensen, J. F., Hollesen, J., Meldgaard, M., Bunce, M., & Hansen, A. J. (2022). Ancient DNA provides insights into 4,000 years of resource economy across Greenland. Nature Human Behaviour, 6(12), Article 12. https://doi.org/10.1038/s41562-022-01454-z
Severson, A. L., Byrd, B. F., Mallott, E. K., Owings, A. C., DeGiorgio, M., de Flamingh, A., Nijmeh, C., Arellano, M. V., Leventhal, A., Rosenberg, N. A., & Malhi, R. S. (2022). Ancient and modern genomics of the Ohlone Indigenous population of California. Proceedings of the National Academy of Sciences, 119(13), e2111533119. https://doi.org/10.1073/pnas.2111533119
Skov, L., Peyrégne, S., Popli, D., Iasi, L. N. M., Devièse, T., Slon, V., Zavala, E. I., Hajdinjak, M., Sümer, A. P., Grote, S., Bossoms Mesa, A., López Herráez, D., Nickel, B., Nagel, S., Richter, J., Essel, E., Gansauge, M., Schmidt, A., Korlević, P., … Peter, B. M. (2022). Genetic insights into the social organization of Neanderthals. Nature, 610(7932), Article 7932. https://doi.org/10.1038/s41586-022-05283-y
ter Schure, A. T. M., Bruch, A. A., Kandel, A. W., Gasparyan, B., Bussmann, R. W., Brysting, A. K., de Boer, H. J., & Boessenkool, S. (2022). Sedimentary ancient DNA metabarcoding as a tool for assessing prehistoric plant use at the Upper Paleolithic cave site Aghitu-3, Armenia. Journal of Human Evolution, 172, 103258. https://doi.org/10.1016/j.jhevol.2022.103258
Vespasiani, D. M., Jacobs, G. S., Cook, L. E., Brucato, N., Leavesley, M., Kinipi, C., Ricaut, F.-X., Cox, M. P., & Romero, I. G. (2022). Denisovan introgression has shaped the immune system of present-day Papuans. PLOS Genetics, 18(12), e1010470. https://doi.org/10.1371/journal.pgen.1010470
John Hawks Newsletter
Join the newsletter to receive the latest updates in your inbox.