When did humankind's last common ancestor live? A surprisingly short time ago
The lines of genealogy of living people converge quickly into the past. Our last genealogical common ancestor lived within the last few thousand years.
Back in 2001, I was a postdoc working on human genomics when Steve Olson talked with me for an interview. Steve was doing background research for a book about how geneticists were changing our understanding of human history. He was excited about some recent work in applied math. Today's humans must all share a common ancestor within the last 5000 years, he explained.
Everyone? I asked. What about the people who came to the Americas more than 12,000 years ago?
Everyone, he said, from Aboriginal people in Australia to Tierra del Fuego. It was a simple consequence of math.
The logic was that in each generation the potential number of ancestral lines doubles. Two parents, four grandparents, eight great-grandparents, and so on. In 20 generations, that makes a million possible lines of ancestry; in 40 generations a trillion.
A trillion lines of genealogy is impossibly large, way larger than the number of humans that existed. These potential lines cannot have been separate; they must converge to a smaller number of actual ancestors. Within a surprisingly small number of generations—the base 2 logarithm of the population size, in fact—one of those actual ancestors will turn up in everybody's family tree. For a million people in one well-mixed population, their most recent common ancestor should have lived around 20 generations ago.
I wasn't prepared for that leap. At the time I was running a lot of simulations with gene genealogies, and I had gotten used to thinking from the gene's point of view. This required looking backward in time in a counterintuitive way.
For example, you have two biological parents and you carry two copies of most of your genes, one inherited from each parent. But you don't carry four copies from your four grandparents. Your father's sperm and mother's egg were products of meiotic cell divisions, leaving them with only one of each chromosome instead of two. You inherited a copy of each gene from two of your grandparents, not four. Those two copies came from two of your great-grandparents, not eight, and from two of your great-great-great-great-great-great grandparents, not 256. It's always two genetic ancestors, no matter how large the genealogy actually was.
Until, that is, those two ancestors coalesce back into one. All of us are distant cousins, and so were all of our parents. Each of us is inbred, it's just that most of us don't know who those shared relatives were. Stepping back in time, generation by generation, your two copies of each gene trace back to a single ancestor. Knowing when this common genetic ancestor lived can reveal a lot—not so much about your immediate family, but about how the population evolved. A geneticist can extend the same idea to more and more people, taking their shared genetic ancestors step by step, to trace the most recent common ancestor, or MRCA, of all the copies of the gene.
Limited as I was to the computing power of a 2001 desktop, the gene's perspective on genealogy was nothing short of magic. The coalescent approach meant that you could trace just the ancestors of a single gene in a small sample of people, ignoring all the rest of the thousands or millions of people in a population.
Of course the view I just described is incomplete. A gene is a part of a chromosome. Before meiosis tosses out one of each pair of chromosomes to make the half-genome of a sperm or an egg, the two parental chromosomes recombine with each other. The chromosome you inherit from your father includes some parts from both your paternal grandparents, and your mother's combines your two maternal grandparents. Because of this crossing over, parts of a single DNA sequence sometimes come from two different ancestors, not one. My work was centered on understanding how to use that complication to better understand what had happened to genes in the past.
What I hadn't been imagining was the case where every lineage doubles in every generation. That's the perspective that Olson was thinking about, as he talked with researchers who had begun to investigate genealogy from the individual point of view instead of the genetic point of view. It was this kind of thinking that led to the provocative conclusion that common genealogical ancestors of all of humanity might have lived only a few thousand years ago.
The groundwork for the idea was lain in 1999 by the statistician Joseph Chang. At the time, anthropologists were very excited that the mitochondrial MRCA of humans lived only 100,000 to 200,000 years ago. People inherit their mitochondrial DNA from their mothers, meaning that the mtDNA point of view is the same as the maternal genealogy. Chang realized that the maternal genealogical lineage was only one out of multitudes of genealogical connections, many of which must lead to even more recent common ancestors than the mtDNA.
“Our MRCA is more recent, since the paths from ancestors to descendants consist of all potential paths for genes to be transmitted, and may include paths that did not happen to be taken by any genes.”—Joseph Chang
Chang worked out a little math. In its details it gets a little mathy, but it's not too hard to explain the gist. If you consider your own parents, grandparents, and so on, your genealogical ancestors double with each generation backward in time. How far do you have to count back until your genealogical lines were as numerous as the people in the entire population? In 10 generations, you have 210 lines of ancestry: that's 1024 lines. In 30 generations, you have 230, which is more than a billion. We can ask the simple question, how many generations does it take to have as many lines of ancestry as there were people in the population? The answer, if we take N as the number of people in the population, is log2N generations into the past.
It's not just you. Everybody else living now had N lines of genealogy going back to that time, too. It's pretty hard for everyone to trace back as many genealogical lines as there were people in the whole population and not share one of those ancestors in common.
Not all of those N lines can lead to different people. Even a mathematically ideal random-mating population has inbreeding, so that your genealogical lines converge to somewhat fewer than N ancestors. But keep counting. One more generation, log2N + 1, and you've got 2N genealogical lines. Another generation back, log2N + 2 , makes 4N lines. All these lines weave back into the population of only N people. The genealogies can't stay separate. Within just a few generations around log2N, somebody must have been a common ancestor of everybody.
The real innovation that Chang made wasn't to show that the genealogies must converge, but to prove mathematically that the larger the population, the closer this MRCA will be to log2N generations. Any small population can be a few generations out of whack, by luck of the genealogical draw. But as populations get really big, it becomes very unlikely for the MRCA to be much older or younger than log2N. A population of a million people should have an MRCA around 20 generations ago. If Earth's current population of eight billion people should manage to stay so large for the next 33 generations, roughly a thousand years, then at least one person living now will have become a common ancestor to our entire future species.
“The major problem in applying these results to human populations is that mating is not random in the real world.”—Douglas Rohde, Steve Olson, and Dennis Chang
Despite the beauty of the math, a big hurdle stands in the way of applying it to real people. For more than 40,000 years, our species has not just been transcontinental; we've been transoceanic. Humans began to inhabit Australia and Papua by 50,000 years ago, the Bismarck Archipelago before 30,000 years ago, parts of the Americas before 20,000 years ago, reaching near the southernmost end of South America by 14,000 years ago. The establishment of human populations in each of these places required deep skill and social organization, as well as some luck. It might seem like the MRCA of humans should have lived before human groups began to scatter around the world, not a mere 1000 years ago when they were already long-established in these far-flung regions.
Yet people around the world were not isolated from each other after they were established on different continents. The very fact that people crossed deep water straits, cold steppes, and mountain ranges in the first place shows that they could traverse formidable barriers. Such crossings were not one-time events. Ancient foraging peoples survived through their intimate knowledge of vast landscapes and an ability to forge relationships with neighbors who had different traditions and languages. Anybody who travels from one place to another carries a whole bundle of genealogical connections along for the ride. When they have children, their genealogies begin to intertwine with others in their new homeland, until the ancestors of the immigrants become ancestors of everyone.
To investigate how much the structure of ancient human populations might matter to their MRCA, Steve Olson the writer got together with Chang and the cognitive scientist Douglas Rohde to build computer models of ancient human populations, publishing their results in a 2004 article in Nature. They imagined ancient humans as a network of populations, in which a small fraction of individuals move from their place of birth to join a new population each generation. This population structure did indeed make it longer to the point where the whole global population shared a common ancestor, more than tripling the expected age of the MRCA. But that just means that instead of 1000 years, the time was between 3000 and 4000 years. This is still only 70 to 100 generations ago. By contrast, the last common genetic ancestor of our species lived around 200,000 years ago—50 times longer than the genealogical common ancestor.
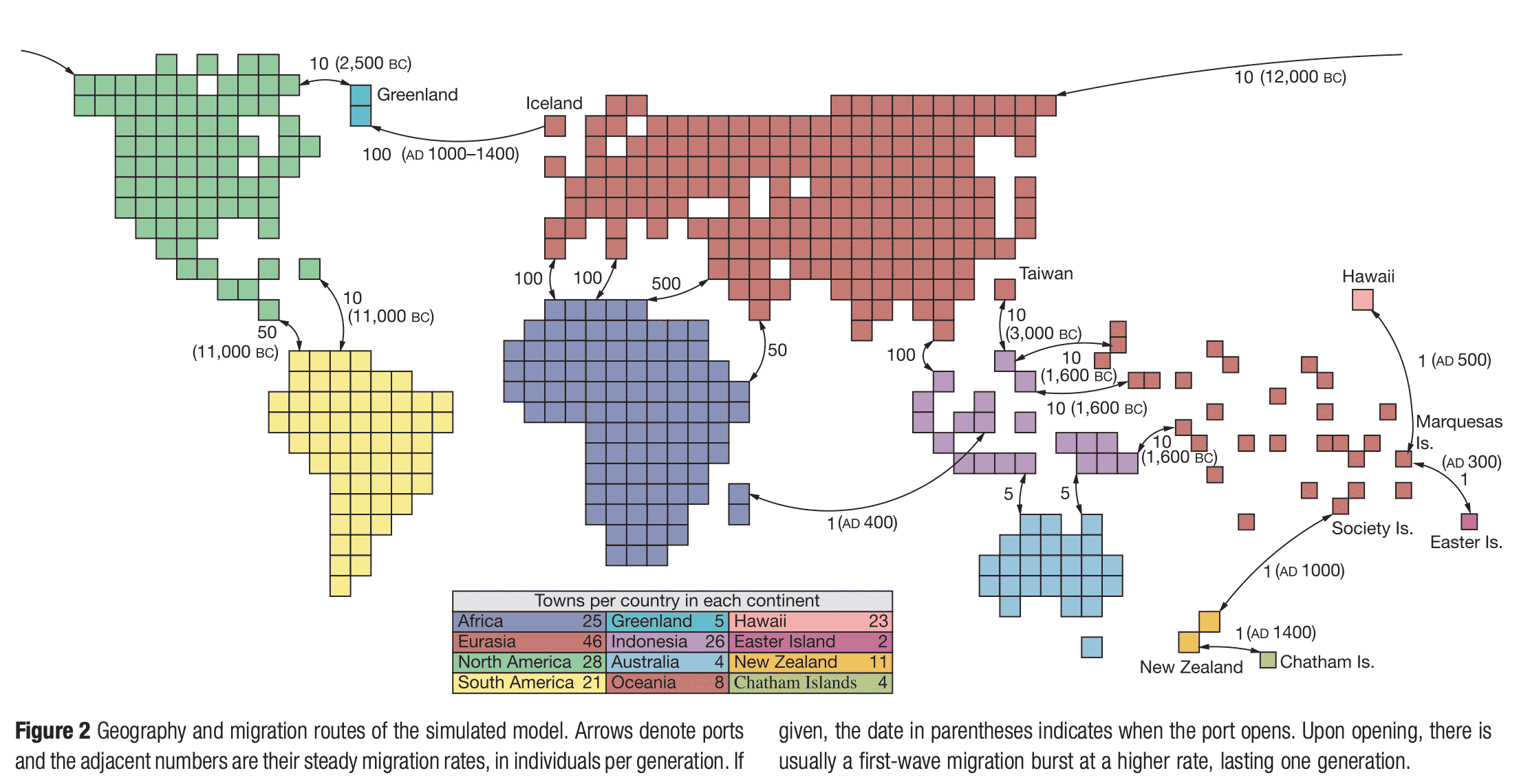
The picture above shows the most detailed of the population models invented by Rohde and coworkers. It's a little like the board game Risk, although with continents segmented into much smaller regions. As intricate as this map may appear, it still takes only around 3400 years for the genealogies of all individuals to point back to a MRCA.
“Actual migration rates among populations are very poorly known and undoubtedly have varied considerably in different times and places.”—Douglas Rohde, Steve Olson, and Dennis Chang
The most important constraints in these models are the links between continents. These merit critical examination. Rohde, Olson, and Chang began with what may seem like a strong claim: “No large group is known to have maintained complete reproductive isolation for extended periods.” They leaned on ethnography and historical records, emphasizing that indigenous peoples have long maintained contacts across the Bering Strait and among the islands of the South Pacific.
The genetic data to examine this claim were not there in 2004. Geneticists at the time were working to understand the first arrival of humans in the Americas, Australia, and many other regions, using genetic information from mtDNA and the Y chromosome. Many geneticists assumed that the mtDNA and Y chromosome haplogroups were markers of ancestral founder populations. The so-called “Seven Daughters of Eve” intepretation of mtDNA in Europe by geneticist Bryan Sykes was one early idea that attributed mtDNA haplogroups to women within a single Upper Paleolithic founding population. Today we recognize that early concept was wrong. Haplogroup diversity in Europe today reflects repeated mixture and expansion of farming and steppe pastoralist populations during the Neolithic and Bronze Age. It took a shift to whole-genome evidence, including ancient DNA, to allow scientists to distinguish mixture and place it in time relative to a population's founding.
We know much more today about the mixture and long-distance contacts among populations during the last few thousand years.
- Ancient DNA has shown the importance of large-scale population expansions and partial replacement of ancestral populations during the last few thousand years. These data have begun to make apparent the biological and genealogical impact of patterns first described by archaeologists or linguists. Along with new understanding of the expansion of Bronze Age steppe populations across Eurasia, geneticists and archaeologists have added new detail to the Bantu expansion across Africa, the Pama-Nyungan language spread across Australia, and the successive spreads of Paleo-Inuit, Inuit, and Yup'ik peoples across the Arctic of North America and Greenland. Human populations were not static once established; they experienced massive immigrant flows across broad geographic regions.
- Genomes from living peoples show how populations were connected across oceans and straits during the mid-to-late Holocene. Examples of this include the contribution of northeast Asian ancestry into present-day speakers of Athabaskan languages in North America, the mid-Holocene infusion of South Asian ancestry into Aboriginal Australian peoples, the increase of Eurasian ancestry among the peoples connected by trading routes along the eastern coast of Africa, and South American ancestry among many of the Polynesian peoples of the South Pacific. Water may have sometimes been a barrier to movement, but during the last few thousand years many groups have used water routes to enable movement and contact across vast distances.
- Archaeologists and geneticists have growing evidence of vast population sizes and trade connections in some parts of the world. A notable example is the prehistory of South America, where large-scale societies flourished across parts of the Amazon basin, and trade networks along the western coast enabled the dispersal of domesticated plants to and from Mesoamerica repeatedly during the early and mid-Holocene.
These connections between continents throughout the last few thousand years left marks on their gene pools that we can still find today, and the mixture of genealogies must have been vastly greater. The small numbers used by Rohde, Olson, and Chang in their models are probably conservative.
As much as we now know about migration throughout the Holocene, the vast movements of the last 500 years have had the greatest effect on the genealogical connectedness of people around the world. This period of human history is much better known. Many of its chapters have tragedy written within them. Disease and genocide devastated many indigenous peoples around the world. The mixture of populations has included the forced assimilation of indigenous people, migration to cities by people deprived of their lands and ways of life, enslavement, human trafficking, and rape. Our intertwining of genealogies is not only a measure of connectedness of populations; it is a witness to legacies that were partially lost.
One of these was the indigenous population of Tasmania. Rising sea levels some 12,000 years ago flooded the land connecting Tasmania to continental Australia, creating the Bass Strait. From this time forward there is no evidence for crossings of the strait until European ships arrived in the 1600s, although archaeologists cannot exclude such crossings for certain. I keep in mind the exceptional sea voyages of Polynesian and Indonesian peoples, and I expect we will someday learn that Tasmania was within their reach. Still, out of all parts of the world where humans lived, Tasmania presents strongest case for isolation for any long period of time.
But the establishment of British colonists in 1803 brought diseases that killed many Aboriginal people, and many others were murdered by colonists or sailors. By the 1820s, the colonial government displaced surviving Aboriginal people into camps, paying bounties for their capture. In these camps disease continued to take a toll. Within a century, all the remaining descendants of Aboriginal Tasmanians were people who also had descended from people of British or other nationalities.
Some indigenous groups today resist contact with outsiders, including those from the nation-states that claim the territories where they live. From the Andaman Islands to the Amazon, there are groups who maintain cultural separation. Their attempts to protect their own cultures are a reaction to the long presence and activities of colonial peoples. But such peoples are not separated from the web of genealogical connections that have surrounded them for thousands of years.
Since the turn of the century, the idea of a recent genealogical ancestor for humankind has made an impact on people's understanding of the past. Olson turned the story into the opening of his 2003 book, Mapping Human History: Genes, Race, and Our Common Origins. Adam Rutherford also discussed the concept in his 2017 book, A Brief History of Everyone Who Ever Lived. Not only scientists, but also religious thinkers have also been inspired by the recent relatedness of humankind. For example, S. Joshua Swamidass used the math to argue that Biblical notions of human descent can live alongside scientific ideas, in his 2019 book, The Genealogical Adam and Eve: The Surprising Science of Universal Ancestry. Many people find comfort in the idea that each of us is part of the same great story, across all ethnicities and races.
This is a lot to carry for a concept that rests on math and not hard data. Archaeologists will never uncover a skeleton that they can identify as the most recent common ancestor of all of us. Indeed, the very idea of a genealogical common ancestor resists attempts to demonstrate with DNA, the most powerful data we can bring to bear about relationships. As special as this person is within our genealogy, it is very likely that most living people have inherited no DNA from this person at all.
This may seem like a paradox: a genealogical ancestor of everybody, from whom most of us have inherited no DNA. It reminds us that genetic and genealogical relationships are different from each other. Many close genealogical relatives are nonetheless genetically and culturally very different from each other. Fifth cousins are not far apart genealogically, but they sometimes share no DNA from their common genealogical ancestors at all.
A similar paradox can be found in our examination of human diversity. Humans are in some ways the least variable of species. And yet, the pattern of genetic diversity across populations of the world marks a complex history of dispersals and interactions. To probe these pattterns, we need to understand a second concept about genealogy: the point at which all members of a population have not only a common ancestor, but exactly the same ancestors.
That will require a second post.
Notes: In addition to the MRCA, another important concept is the identical ancestors point (IAP), the time when all past individuals are either ancestors of everyone living today or no one. In a second post, I will give more detail on this concept and its relationship to ancient introgression.
Two small groups of researchers explored the mathematical properties of the most recent common genealogical ancestor and identical ancestors point in a series of papers early in the 2000s. Manrubia, Derrida, and Zanette provided an accessible account of their work in a 2003 article in American Scientist. Chang, Rohde, and Olson's 2004 Nature article is also highly readable.
A number of people have written popularized accounts of these genealogical concepts. Graham Coop wrote a series starting with “Our vast shared family tree”. Scott Hershberger wrote about the concepts for Scientific American in 2020, as did Adam Rutherford for The Guardian in 2015. Rutherford returned to the topic again for Nautilus, in an excerpt from his book.
References
Chang, Joseph T. Recent common ancestors of all present-day individuals. Advances in Applied Probability 31 (1999): 1002-1026. https://doi.org/10.1239/aap/1029955256
Derrida, Bernard, Susanna C. Manrubia, and Damián H. Zanette. Statistical properties of genealogical trees. Physical Review Letters 82 (1999): 1987. https://doi.org/10.1103/PhysRevLett.82.1987
Derrida, Bernard, Susanna C. Manrubia, and Damián H. Zanette. On the genealogy of a population of biparental individuals. Journal of Theoretical Biology 203 (2000): 303-315. https://doi.org/10.1006/jtbi.2000.1095
Manrubia, Susanna C., Bernard H. Derrida, and Damián H. Zanette. Genealogy in the era of genomics: Models of cultural and family traits reveal human homogeneity and stand conventional beliefs about ancestry on their head. American Scientist 91 (2003): 158-165.
Pugach, I., Delfin, F., Gunnarsdóttir, E., Kayser, M., & Stoneking, M. (2013). Genome-wide data substantiate Holocene gene flow from India to Australia. Proceedings of the National Academy of Sciences, 110(5), 1803-1808. https://doi.org/10.1073/pnas.1211927110
Rohde, Douglas L. T., Steve Olson, and Joseph T. Chang. Modelling the recent common ancestry of all living humans. Nature 431 (2004): 562-566. https://doi.org/10.1038/nature02842
John Hawks Newsletter
Join the newsletter to receive the latest updates in your inbox.