Denisovan traits bring up the old problem of understanding morphological continuity
A paper by Shara Bailey and coworkers suggests that three-rooted lower molars are diagnostic of population mixture from Denisovans.

During the 1980s and 1990s, the idea of multiregional evolution of modern humans was based upon the observation that today’s people living in various regions of the world share traits with archaic humans who lived in the same regions. Such traits provided evidence of regional continuity of ancestry.
This summer a brief report by Shara Bailey, Jean-Jacques Hublin, and Susan Antón suggested that three-rooted mandibular premolars in today’s Asian populations have actually been inherited from Denisovans: “Rare dental trait provides morphological evidence of archaic introgression in Asian fossil record”.
According to this idea, three-rooted mandibular molars are a regional continuity trait in Asia. Molars in the mandibular dentition usually have two roots, but a three-rooted form occurs in East Asian people fairly commonly.
It has long been thought that the prevalence of 3-rooted lower molars in Asia is a relatively late acquisition occurring well after the origin and dispersal of H. sapiens. However, the presence of a 3-rooted lower second molar in this 160,000-y-old fossil hominin suggests greater antiquity for the trait. Importantly, it also provides morphological evidence of a strong link between archaic and recent Asian H. sapienspopulations. This link provides compelling evidence that modern Asian lineages acquired the 3-rooted lower molar via introgression from Denisovans.
Here is the illustration including the virtual model of the Xiahe M2.
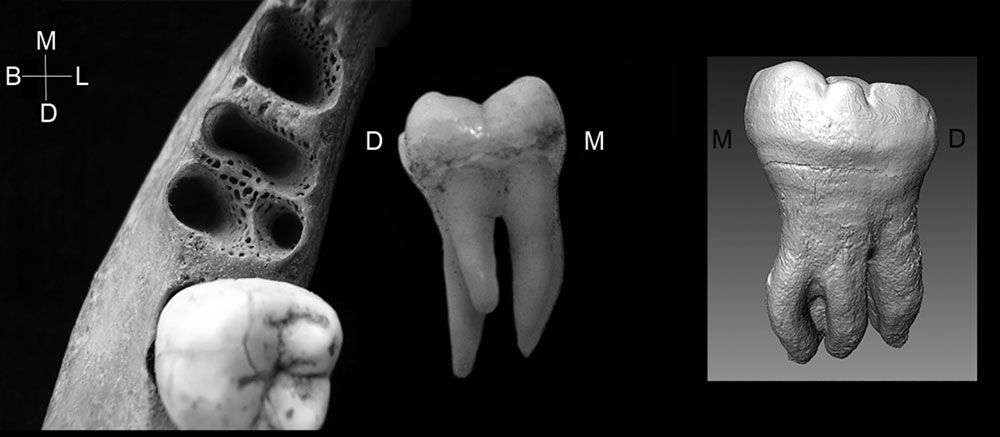
I think that Bailey and coworkers here present an interesting hypothesis. This idea brings back to my mind some of the 1990s-era discussion about how scientists should recognize evidence of genetic admixture or introgression from morphological traits.
During the 1990s, several scientists were highly critical of others’ identification of “continuity features”. They noted that it was difficult to prove that any feature was a unique markers of descent from local archaic humans, because the fossil record is incomplete and biased. Some features that had been identified as continuity traits in Neandertals and later Europeans, they argued, might also have been present in earlier African populations but not yet sampled in the tiny fossil record of the Middle Pleistocene in Africa. Much discussion about regional continuity during the 1990s (and to some extent even earlier) focused upon whether “Neandertal” traits in later Europeans really were uniquely found in Neandertals, or whether they might have been in other populations at non-negligible frequencies.
Many advocates of an “out of Africa” model of evolution also suggested that apparent continuity traits might instead have resulted from parallelism or convergence in the newly-arrived modern humans. In their view, early modern humans might rapidly evolve to become similar to archaic humans in the same parts of the world, because of natural selection in the same local environments.
My own concern at the time was that continuity traits suffer an analytical bias that is hard to deal with. A strange trait that we see in a Neandertal is easy to notice when we see it in later European skeletal remains. But we don’t really know how likely such similarity may be without considering the total number of traits we didn’t notice.
This brings us back to sampling. In the period just before the apparent dispersal of modern humans from Africa, anthropologists have found many Neandertals and relatively few of anything else. If we accept that the Xiahe mandible is really a Denisovan (which maybe we should be more cautious about), that still brings the net number of Denisovan specimens with lower first molars to the grand total of one.
I always thought it was quite reasonable to start from the assumption that the data probably reflect a simple mixture model, and a Neandertal trait that was present at a lower frequency in Upper Paleolithic Europeans probably reflected some ancestry into that population from Neandertals.
I still think that’s reasonable, but my perspective has changed a bit. The main difference in my thinking today is that we know that the fractions of ancestry from many archaic groups are small—on the order of a few percent. I don’t think we can predict much about how such small levels of ancestry may affect morphology.
Europeans do have Neandertal ancestors—we now know that beyond any question. But East Asians today also have Neandertal ancestors. In fact, Neandertals contributed more genetic ancestry to East Asians than they did to Europeans. And Neandertals contributed more than ten times as much ancestry to East Asians as Denisovans contributed to East Asians.
Which morphological traits of East Asians today are markers of their Neandertal ancestry? That’s a question that presently has no answer. The answer might even be “none”. But it would be strange for East Asian people today to have no skeletal signs of Neandertal ancestry if European people (with less Neandertal ancestry) have some skeletal features that point that direction.
And it would be even more remarkable for many East Asians to have a dental trait that uniquely reflects their 0.002 fraction of Denisovan ancestry and have none that reflect their 0.02 fraction of Neandertal ancestry.
Maybe it’s really true. If so, we should probably guess that natural selection has acted on three-rooted mandibular molars differently from other traits, favoring its persistence in later populations.
I think it’s good to be alert for morphological connections between living and archaic groups. I don’t assume that they will reflect the frequency of contribution of genes from archaic groups. If the frequency of a trait diverges a great deal from the small fraction of genetic ancestry, it may be a sign of interesting evolutionary dynamics.
Reference
Bailey, S. E., Hublin, J.-J., & Antón, S. C. (2019). Rare dental trait provides morphological evidence of archaic introgression in Asian fossil record. Proceedings of the National Academy of Sciences, 116(30), 14806–14807. https://doi.org/10.1073/pnas.1907557116
John Hawks Newsletter
Join the newsletter to receive the latest updates in your inbox.



