The real story of myosin, jaw muscles, and ancient brains
The provocative idea that our genus arose with a deactivated muscle gene turned out to be wrong.

Back in 2004, I was excited to read the new story of an ancient genetic change that might explain anatomical changes in fossil hominins. Hansell Stedman and collaborators at the University of Pennsylvania had collected sequence data for a gene called MYH16. Previous work showed that this gene had lost its normal function in humans, making it a pseudogene. Other primates express this gene in their jaw muscles, where it strengthens the bite force. By comparing the sequences of humans and other primates, Stedman's team estimated that MYH16 had lost its function in human ancestors around 2.4 million years ago. Maybe, the researchers suggested, this genetic change caused the evolution of smaller jaws and teeth in our genus, Homo.
It was just a study of a single gene, but this work generated an outsized reaction. It was the first time that anyone had identified a specific gene that caused a change we see in fossil hominins.
Today we know that the crucial result of this study—the timing of the MYH16 deactivation—was wrong. This gene has nothing to do with the appearance of Homo; in fact, even fossil hominins with the largest teeth and jaws probably shared this genetic change with us. Yet a steady stream of books and articles about human origins are still repeating the idea, nearly twenty years later.
The real story is not so simple, but no less interesting. There's no question that the timing was all wrong. The change in jaw muscles happened long before our genus arose. What's fascinating is that the big jaws of many of our fossil ancestors weren't built for nutcracking power. Meanwhile, gene expression data suggest that the MYH16 gene isn't as dead as it looks.
The original story
The protein product of MYH16 is one of the heavy chain myosins, a kind of protein that works with actin to enable muscle fibers to contract. The protein is denoted as MyHC-M, for myosin heavy chain–masticatory. In most vertebrates this protein is expressed in muscles that develop from the first pharyngeal arch, including temporalis, which in mammals is the largest muscle of the jaw. But humans do not produce the MyHC-M in these muscles. The human coding sequence of MYH16 has a frameshift mutation that prevents translation of the functional product.
Stedman and coworkers set out to discover when this gene lost its function. They looked at more than 1100 base pairs of sequence from a chimpanzee, orangutan, macaque, and a dog, and they found that the coding sequence in humans has a small number of unexpected amino-acid-coding changes—changes that wouldn't be likely if the gene product was still being maintained by selection. From this small number of new changes, they estimated when the gene lost its function: 2.4 million years, to which they applied an error factor plus or minus 300,000 years.
To these researchers, the estimate held meaning because of its correspondence with early evidence of fossil Homo, which had smaller jaw muscles than earlier hominins. In the last sentence of their discussion, Stedman and colleagues advanced a provocative hypothesis: Not only did Homo have smaller jaws and teeth, but reducing the jaw muscles may have enabled the evolution of larger brain size.
“Our findings on the age of the inactivating mutation in the MYH16 gene raise the intriguing possibility that the decrement in masticatory muscle size removed an evolutionary constraint on encephalization, as suggested by the anatomy of the muscle attachments relative to the sutures.”—Hansell Stedman and coworkers
In this scenario, mechanical forces on the skull once held back the evolution of brain size in early hominins. When those forces were relaxed by reducing the jaw musculature, brain size was free to increase.
Genes and morphology
Looking back at the study today, I think probably many people may wonder how it made such a splash. Even at the time, evidence showed that large-brained Homo actually had thicker vault bones compared to big-jawed Australopithecus and Paranthropus. In a 2006 essay, Melanie McCollum and collaborators would point out that children grow adult-sized brains at a much younger age than adult-sized jaw muscles, making the idea of a mechanical contraint on brain evolution fail to add up.
I remember when the study came out, I brought it in to discuss with my undergraduate students in human evolution. At that time, geneticists knew of some human genes where coding mutations could cause Mendelian disorders that affect morphology. But every one of these conditions was rare, and examination of these genes in other primates had not yet found any evidence that they were involved in morphological evolution in our lineage.
Evolutionary geneticists were debating whether protein-coding changes were likely to explain morphological evolution at all. They pointed to the pioneering work of Mary-Claire King and Allan Wilson, who noted that humans and chimpanzees are so genetically similar that differences in proteins are unlikely to explain their morphological and behavioral differences. Instead, they argued, changes in the regulation of genes must be responsible for most of these biological differences.
“A relatively small number of genetic changes in systems controlling the expression of genes may account for the major organismal differences between humans and chimpanzees.”—Mary-Claire King and Allan Wilson
Such biologists found their work aligned with the growing field of evolutionary developmental biology, known as “evo-devo”. They studied mutant strains of mice, fruit flies, and other laboratory organisms to understand how changes to gene expression make a difference to the development of morphology. These experiments were showing that scientists could cause large morphological changes in an organism by turning genes on or off at critical times.
These experimental models revived an old idea in human evolution, that instead of gradual evolutionary trends, some morphological innovations might have happened in a few large steps. Pseudogenes were a natural place to look: natural versions of the gene knockouts that developmental geneticists were creating in their laboratories.
Developmental biologists leaped to promote the potential importance of the MYH16 discovery. Nature commissioned the developmental biologist Pete Currie to write a commentary to accompany the paper by Stedman and coworkers. Currie opened with a reference to the historic debate between Thomas Huxley and Bishop Wilberforce over the validity of Darwin's ideas, then dramatically turned to the MYH16 research:
“In the history of this debate, the paper by Stedman et al. is of special significance. It describes what may be the first functional genetic difference between humans and apes.”—Pete Currie
Another evo-devo scientist, Sean B. Carroll, featured the MYH16 work in his best-selling book, Endless Forms Most Beautiful. Explaining that “muscle anatomy has a large effect on bone growth”, Carroll recounted that smaller jaw muscles might indeed have changed the evolution of the braincase, allowing it to become “thinner and larger”. He went a step further. Noting that spoken language requires fine motor control of the jaw, Carroll suggested that the loss of MyHC-M may have faciliated the evolution of speech.
The story unravels
Within a little over a year after its publication, key parts of the story were proven wrong. George Perry, Brian Verrelli, and Anne Stone took a closer look at the sequence of MYH16, expanding the data to include more than 30 kilobases of coding and noncoding DNA. This larger dataset yielded very different results from the more limited data that Stedman and coworkers had used.
When Perry, Verrelli, and Stone looked downstream of the frameshift mutation, they found many additional amino-acid-coding changes. This expanded number made the age of the gene's deactivitation look much older. Instead of 2.4 million years ago, it seemed that these downstream parts of MYH16 had lost function more than 5 million years ago. Whatever had happened to human jaw muscles was not just in the genus Homo, it would also have been shared with hominins that had much bigger jaws and teeth, Australopithecus and Paranthropus. The idea that the functional loss of this gene could explain the reduction of jaw muscles in Homo was simply wrong.
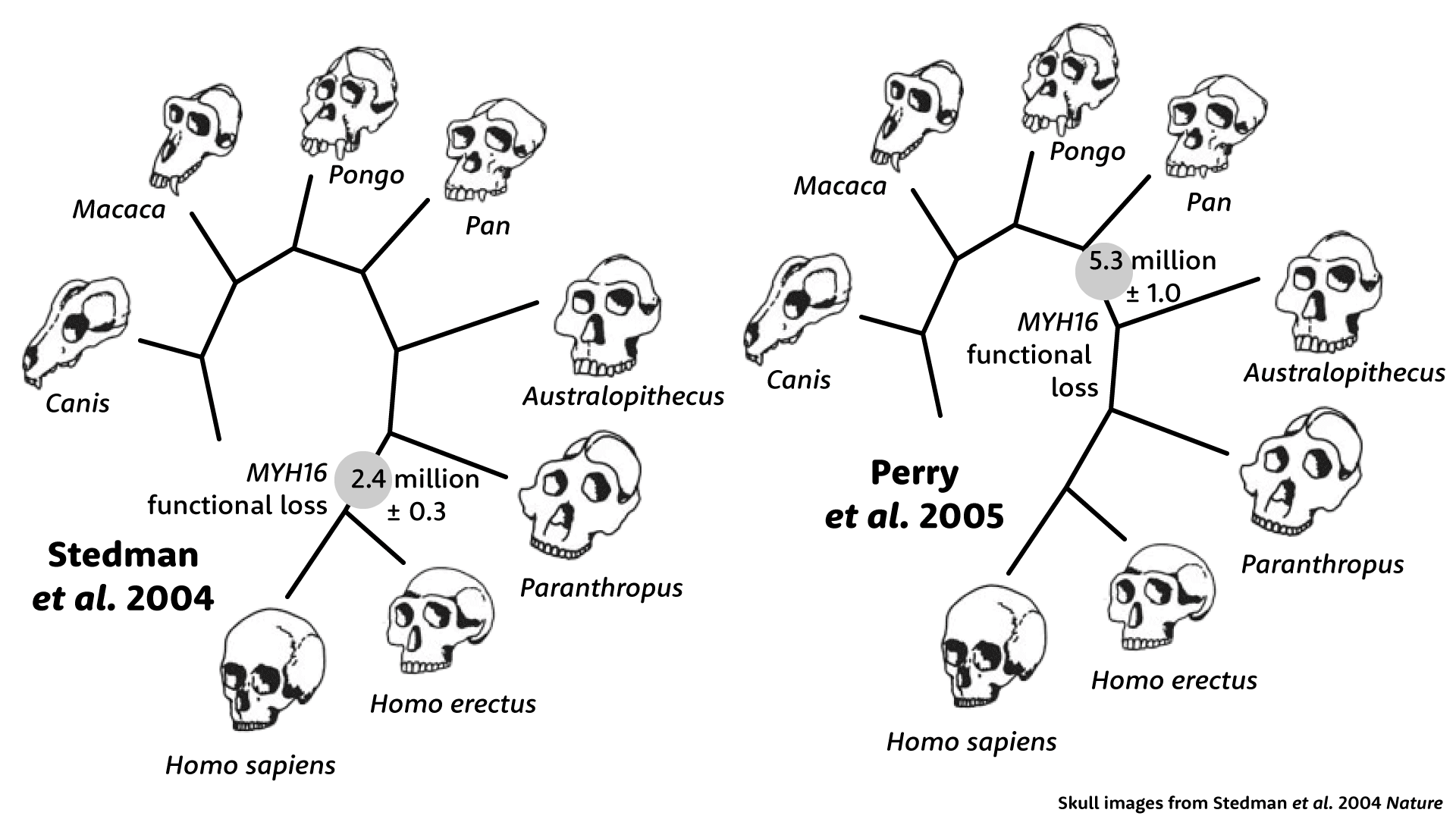
Perry and coworkers found something even more surprising. Upstream of the frameshift, the coding sequence of MYH16 shows a very different picture than downstream. Upstream, the sequence looks almost exactly like other primates, with hardly any amino-acid-coding substitutions. This part of the gene had evolved as if it were still under selection. But how was that possible, if the gene had lost its function? Nobody could say.
Later work has shown that MYH16 is transcribed into mRNA not only in the muscles of the head but in many parts of the human body. In a 2013 survey of protein fragments in a human cell line, Rui Branca and coworkers found that the downstream portion of MYH16 was being translated into protein, a shorter form than the MyHC-M in other primates. In 2019, Yi Shao and collaborators found that both the upstream and downstream portions of the gene match expressed sequence tags from ribosomes. So far, the only known biological correlate of MYH16 expression in humans is negative: the gene is upregulated in some cancers, and Ming Sun and coworkers found that in pancreatic cancer MYH16 expression was negatively correlated with survival. But the area of chromosome 7 around MYH16 shows statistical evidence that a haplotype has increased in frequency over the past several thousand years. Some functional role for this gene may be waiting to be discovered.
Jaws and muscles
More is understood today about the function of the MyHC-M protein in the jaws of species that have it. The temporalis includes both type I (slow) and type II (fast) muscle fibers as well as fibers that are a hybrid of both types, and each type can vary in myosin isoform composition. The fast myosin isoforms in type II fibers generate high power at the cost of fatigue, and those with MyHC-M are a bit slower but exert the strongest pull. Jaw muscle fibers also can include MyHC-α-cardiac, the heavy chain myosin most expressed in heart muscle, which is intermediate between type I and type II in contractile force while resisting fatigue like type I fibers.
In 2004, a lot was already known about the histology of temporalis muscle fibers and the presence of MyHC-M in many species. Stedman and coworkers added to this research, including powerful images showing a section of human temporalis muscle tissue with much smaller type II fibers than found in macaque temporalis. The idea was that the deactivation of the gene might have caused an immediate reduction in muscle size.
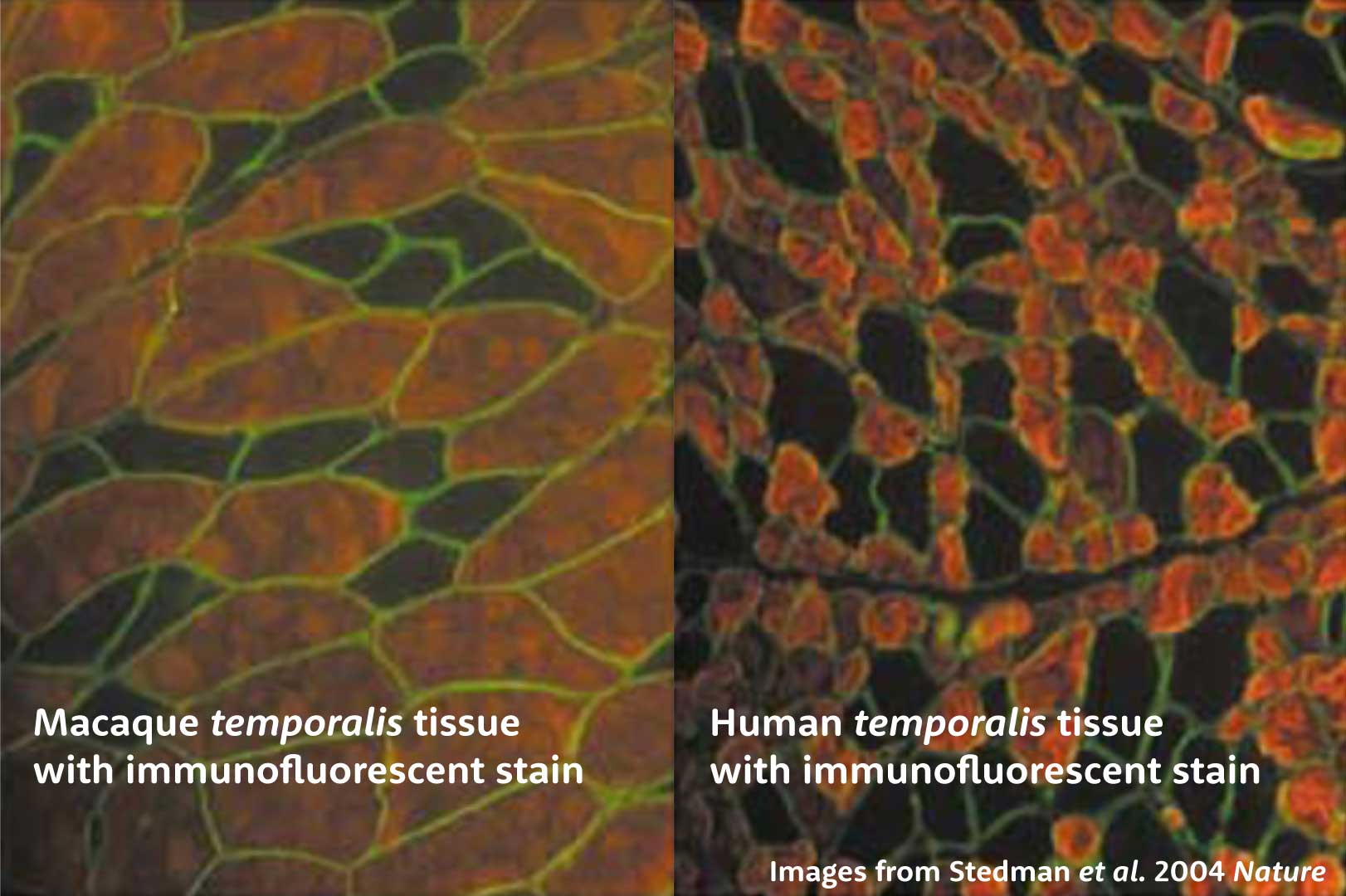
But I find that one of the drawbacks of many research papers in this area is that they portray only one or a handful of comparisons, when the temporalis muscle is a large and heterogeneous structure. In the last decade or so, some researchers have tried to get a better idea of the way that the different kinds of myosins contribute to overall force production in the muscle.
For example, Megan Holmes and Andrea Taylor recently quantified the proportions of fibers with different myosin isoforms in the jaw muscles of several species of nonhuman primates. In this paper they showed sections of the temporalis muscle of three species—tufted capuchins, crab-eating macaques, and western gorillas—representing three of the major groups of living primates. For each section, they made four very thin slices, treating each with a different stain that binds to specific myosin isoform. This gives a picture of the fibers that have skeletal slow, α-cardiac, skeletal fast, and masticatory heavy chain myosins.
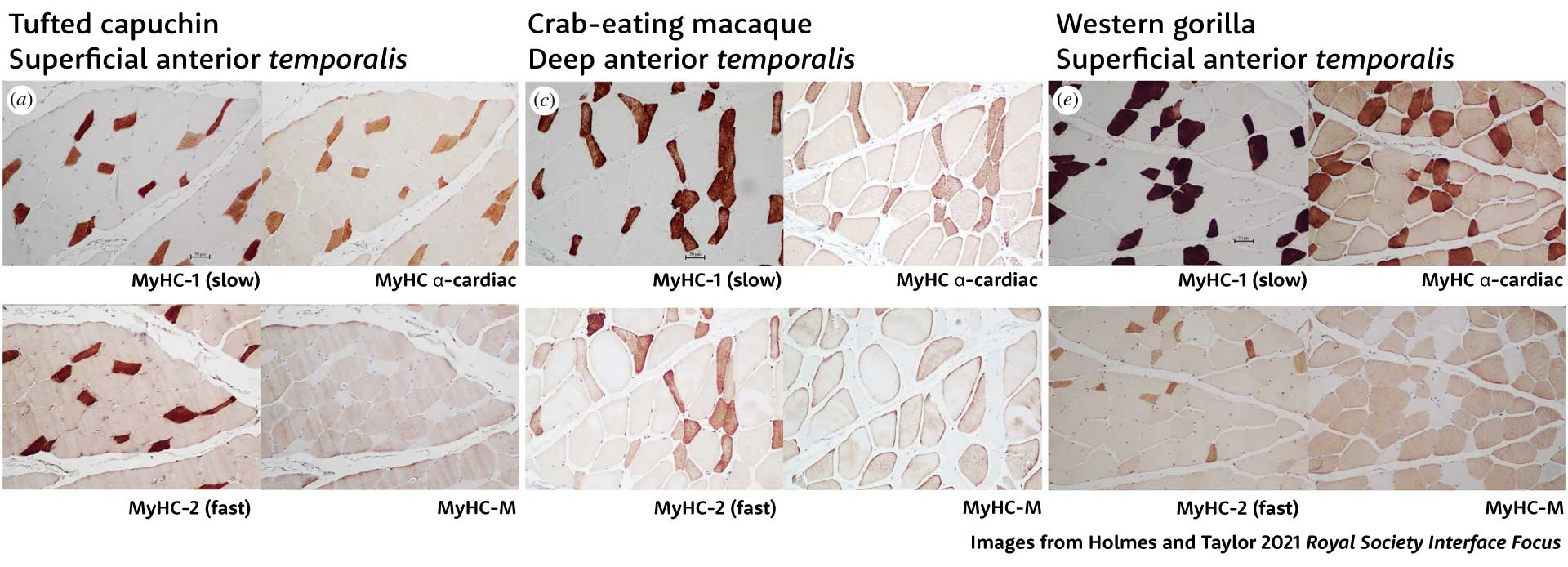
Holmes and Taylor found that most of the fibers in these primates were hybrids, with nearly all either slow fibers that blend MyHC-1 and MyHC-α-cardiac with some fast MyHC-2, or fast fibers with MyHC-2, MyHC-M, and MyHC-α-cardiac.
In another paper, Taylor and Holmes studied the jaw muscles of four gorillas, four chimpanzees, and two bonobos. When they looked at the different parts of the temporalis muscle, they found some big contrasts in the cross-sectional areas of fiber types. Chimpanzee jaw muscles have more fast fibers, which include MyHC-M, than gorillas or bonobos. Compared to the superficial part of the muscle, the deep anterior temporalis has more slow fibers in all three species. The most striking finding was that 96% of the gorilla deep temporalis is made up of slow fibers.
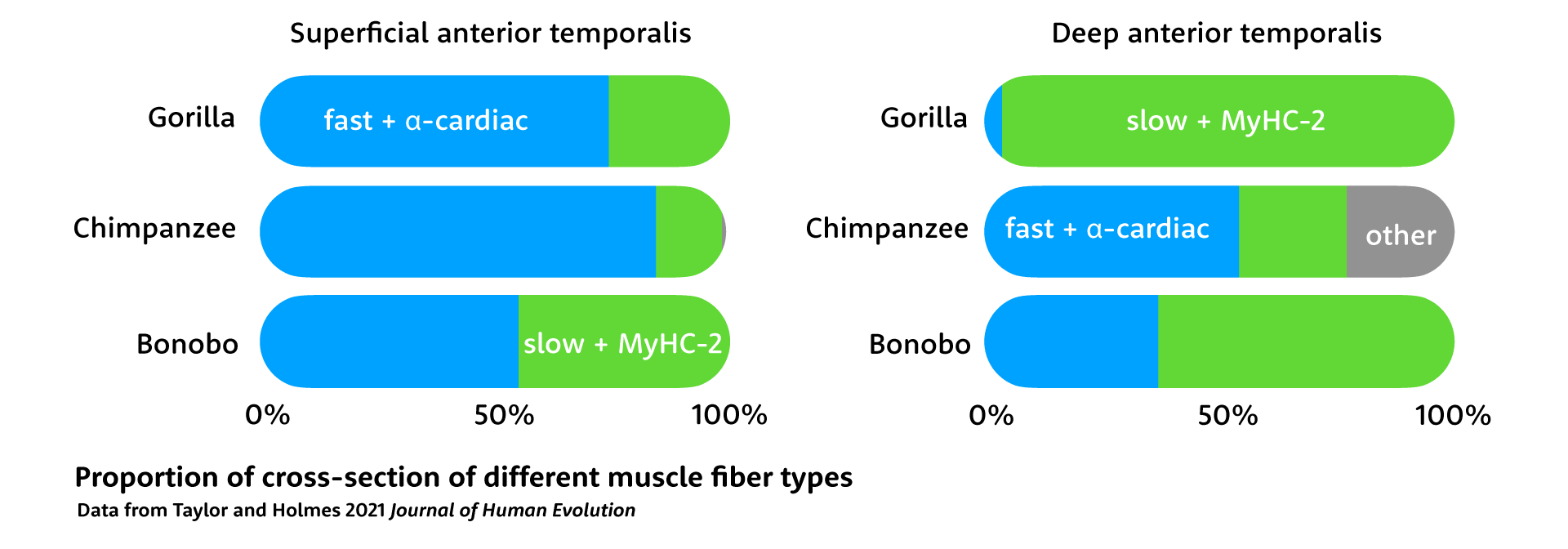
These differences in fiber composition make sense considering how these apes use their jaw muscles. Gorillas spend more time feeding, consuming more leaves and stems and less fruit. They chew a lot more than chimpanzees or bonobos, and gain greater chewing endurance from the high proportion of slow fibers, as well as the inclusion of the α-cardiac isoform in their fast fibers. Gorilla jaw muscles are much larger than the other African apes, but Taylor and Holmes point out that they are less powerful than their larger size would make it seem. When they need rapid power, gorillas can recruit their superficial anterior temporalis, while they can lean on the deep muscle for longer duration chewing.
Gorillas are not unique in accentuating slow fibers in their jaw muscles. Many mammals that rely on grazing or longer-duration chewing likewise have fewer fast fibers. Gorillas still produce the MyHC-M isoform but many grazing mammals, including most grazing ungulates, marsupial grazers, rodents, and rabbits do not express masticatory myosin at all. Humans are not unique; we fall in with species that chew a lot.
The chewing jaws of early hominins
These comparisons suggest that it wasn't wimpy jaw muscles that underlies the loss of masticatory myosin in our lineage. A better hypothesis is that large-jawed ancestors needed a higher fraction of slow fibers for long-duration chewing.
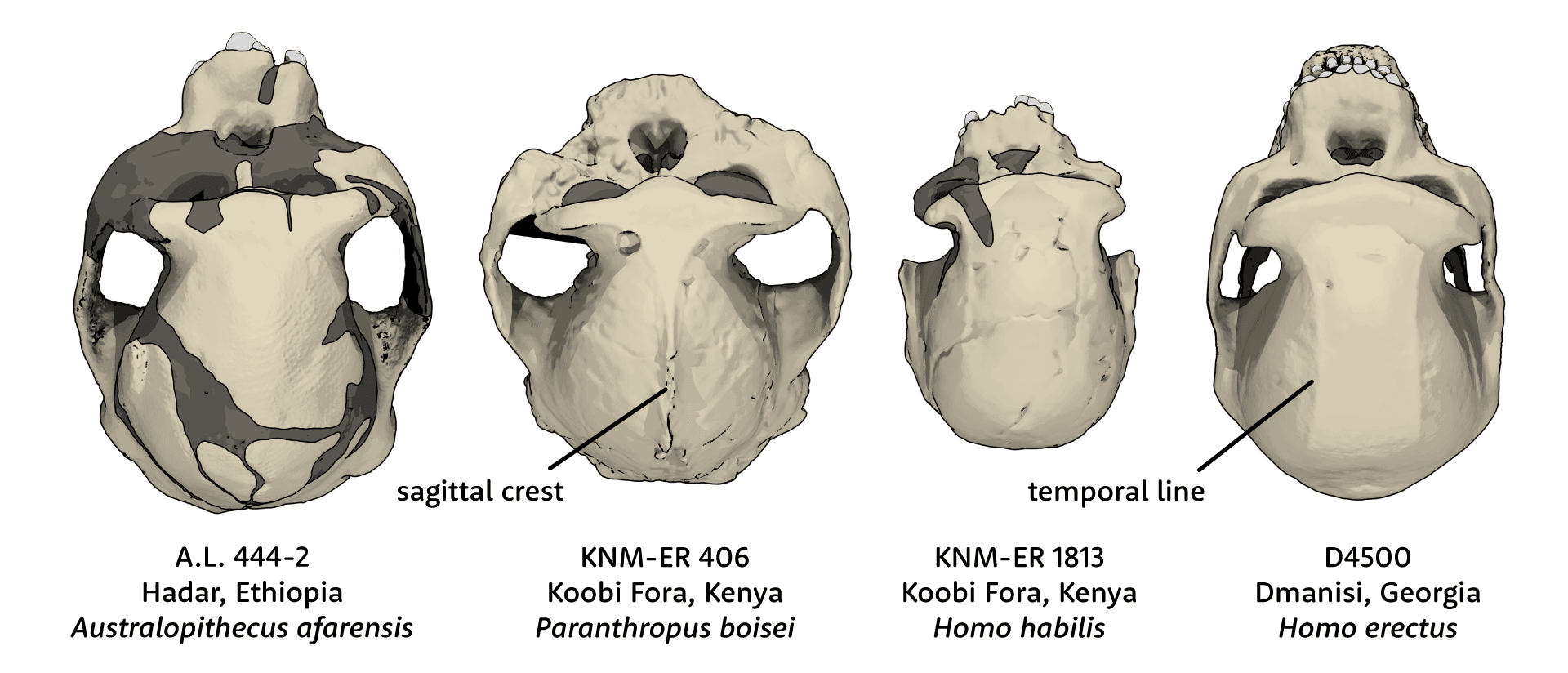
Human ancestors stopped making MyHC-M more than 5 million years ago. Our current understanding of hominin phylogeny suggests that neither Homo, Australopithecus, nor Paranthropus yet had diversified at that early time in our evolution. So it is likely that the MYH16 frameshift mutation happened in a stem ancestor of all these later hominins. Without direct fossil evidence of this ancestor, we cannot say for sure what its jaw muscles or diet may have been like. But most of the early descendants of this stem, including all species of Australopithecus, Kenyanthropus, and Paranthropus, had larger jaw muscles, molars, and premolars than either today's humans or today's chimpanzees and bonobos. Certainly the absence of MyHC-M didn't stop hominins from having large and powerful jaws.
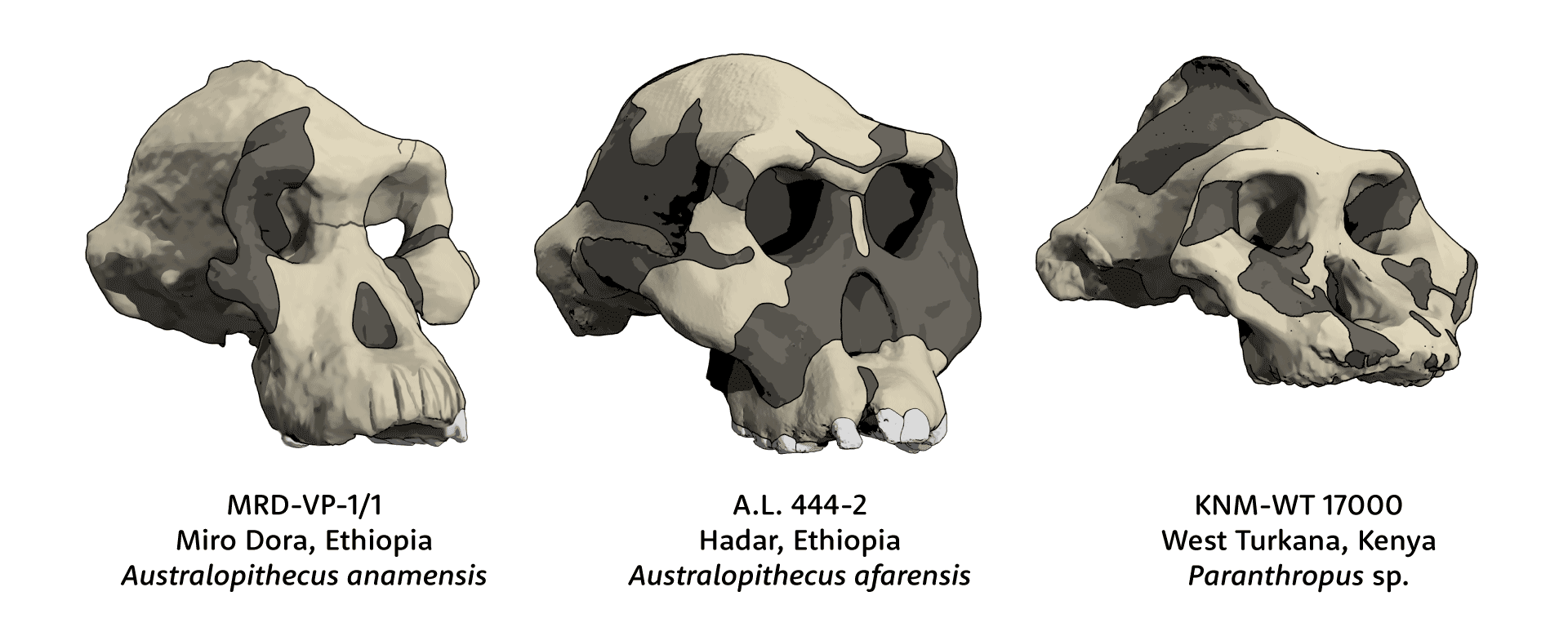
Meanwhile there is growing evidence that early hominins had long feeding times with lots of chewing. The hominin with the largest jaw musculature was Paranthropus boisei. Anthropologists once imagined this species as “Nutcracker Man”, but evidence from dental microwear shows that the species rarely ate hard food items that would have required massive bite force. Instead, the evidence suggests the species ate fibrous plant parts that took time to chew. Jaw muscles with a high proportion of slow fibers, similar to gorillas, would have served this species well. Like many other kinds of mammals with long chewing demands, Paranthropus got by without MyHC-M.
It's true that most fossil Homo after 1.8 million years ago had reduced jaw musculature compared to Australopithecus or Paranthropus. But these overlapped a lot with each other, and most of the earliest fossils attributed to Homo still have markedly thick mandibles and big teeth in the size range of Australopithecus. Besides, as Taylor and Holmes have pointed out, the strength of jaw muscles is not a straightforward result of their size. The robusticity of the mandible, vault, and face of early Homo erectus attests that their temporalis and masseter muscles still had a lot of power. If they chewed less than the big-toothed Paranthropus, which seems likely, their jaw muscles probably had more fast fibers, as today's chimpanzees do. They were just built without masticatory myosin, yielding a little less strength than some other primates, but just as much rapid control.
What remains unknown is whether MYH16 was doing anything valuable besides making masticatory myosin. The frameshift mutation made the translation of MyHC-M impossible for hominins, but left shorter potential translated products both upstream and downstream. The upstream portion continued to evolve under selective constraint, while the downstream portion rapidly changed. If these products have biological roles, then the loss of MyHC-M may have freed them to evolve in distinct ways in our lineage.
MYH16 was once credited with a massive role in our evolutionary story. Popular books and scientific articles in many fields are still citing the story and the date of 2.4 million years to this day. This turned out to be wrong. The real evolution of this gene and the chewing system of hominins is a lot more interesting than the simple hypothesis suggested. Beyond this, there is the potential of an unexpected genetic story still to be revealed.
Notes: This story was fun to investigate and took me in a direction I didn't expect when I started. The possibility that translated products of MYH16 may be functional is a ripe area for research. The gene keeps popping up in genome-wide transcription and expression analyses, without any specific investigation of how the new products work. It's not yet clear how MYH16 matters to the cancers where it is expressed, or whether its expression is just side effect of increased activity of some transcription factors. Nobody knows whether the smaller protein products may have functions in normal tissues, or whether alternative splicing may also produce them in other primates.
During the early 1980s scientists developed a polyclonal antibody test for MyHC-M, enabling them to test for its presence in the muscle tissue of a wide array of vertebrates. The best review of the distribution of MyHC-M across the vertebrate phylogeny is still a 2002 paper by Joseph Hoh. It's remarkable to look back at this research, accomplished long before the MYH16 gene was characterized, to see how histologists developed knowledge of the fiber types in various kinds of muscles.
Did I really label the “Black Skull”, KNM-WT 17000, as Paranthropus sp. instead of P. boisei or P. aethiopicus? You bet I did. With the discovery of Paranthropus as early as 2.9 million years ago at Nyayanga, Kenya, I think we have a lot to learn about the variation and evolution of early forms of this large-toothed hominin. In light of the lack of diagnostic morphology of the Omo 18-18 jaw, I assess P. aethiopicus as a nomen dubium, which can't be compared directly to the KNM-WT 17000 skull.
References
Branca, R. M. M., Orre, L. M., Johansson, H. J., Granholm, V., Huss, M., Pérez-Bercoff, Å., Forshed, J., Käll, L., & Lehtiö, J. (2014). HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nature Methods, 11(1), Article 1. https://doi.org/10.1038/nmeth.2732
Carroll, S. B. (2006). Endless Forms Most Beautiful: The New Science of Evo Devo. W. W. Norton & Company.Hoh, J. F. Y. (2002). `Superfast’ or masticatory myosin and the evolution of jaw-closing muscles of vertebrates. Journal of Experimental Biology, 205(15), 2203–2210. https://doi.org/10.1242/jeb.205.15.2203
Currie, P. (2004). Muscling in on hominid evolution. Nature, 428(6981), Article 6981. https://doi.org/10.1038/428373a
Hoh, J. F. Y. (2002). `Superfast’ or masticatory myosin and the evolution of jaw-closing muscles of vertebrates. Journal of Experimental Biology, 205(15), 2203–2210. https://doi.org/10.1242/jeb.205.15.2203
Holmes, M., & Taylor, A. B. (2021). The influence of jaw-muscle fibre-type phenotypes on estimating maximum muscle and bite forces in primates. Interface Focus, 11(5), 20210009. https://doi.org/10.1098/rsfs.2021.0009
King, M.-C., & Wilson, A. C. (1975). Evolution at Two Levels in Humans and Chimpanzees. Science, 188(4184), 107–116. https://doi.org/10.1126/science.1090005
Koncina, E., & Letellier, E. (2020). Chapter Six - Myosins: Driving us towards novel targets and biomarkers in cancer. In C. Thomas & L. Galluzzi (Eds.), International Review of Cell and Molecular Biology (Vol. 356, pp. 291–322). Academic Press. https://doi.org/10.1016/bs.ircmb.2020.06.004
Lee, L. A., Karabina, A., Broadwell, L. J., & Leinwand, L. A. (2019). The ancient sarcomeric myosins found in specialized muscles. Skeletal Muscle, 9(1), 7. https://doi.org/10.1186/s13395-019-0192-3
McCollum, M. A., Sherwood, C. C., Vinyard, C. J., Lovejoy, C. O., & Schachat, F. (2006). Of muscle-bound crania and human brain evolution: The story behind the MYH16 headlines. Journal of Human Evolution, 50(2), 232–236. https://doi.org/10.1016/j.jhevol.2005.10.003
Perry, G. H., Verrelli, B. C., & Stone, A. C. (2005). Comparative Analyses Reveal a Complex History of Molecular Evolution for Human MYH16. Molecular Biology and Evolution, 22(3), 379–382. https://doi.org/10.1093/molbev/msi004
Schiaffino, S., & Reggiani, C. (2011). Fiber Types in Mammalian Skeletal Muscles. Physiological Reviews, 91(4), 1447–1531. https://doi.org/10.1152/physrev.00031.2010
Shao, Y., Chen, C., Shen, H., He, B. Z., Yu, D., Jiang, S., Zhao, S., Gao, Z., Zhu, Z., Chen, X., Fu, Y., Chen, H., Gao, G., Long, M., & Zhang, Y. E. (2019). GenTree, an integrated resource for analyzing the evolution and function of primate-specific coding genes. Genome Research, 29(4), 682–696. https://doi.org/10.1101/gr.238733.118
Stedman, H. H., Kozyak, B. W., Nelson, A., Thesier, D. M., Su, L. T., Low, D. W., Bridges, C. R., Shrager, J. B., Minugh-Purvis, N., & Mitchell, M. A. (2004). Myosin gene mutation correlates with anatomical changes in the human lineage. Nature, 428(6981), Article 6981. https://doi.org/10.1038/nature02358
Sun, M., Wang, Y., Zheng, C., Wei, Y., Hou, J., Zhang, P., He, W., Lv, X., Ding, Y., Liang, H., Hon, C.-C., Chen, X., Xu, H., & Chen, Y. (2021). Systematic functional interrogation of human pseudogenes using CRISPRi. Genome Biology, 22(1), 240. https://doi.org/10.1186/s13059-021-02464-2
Taylor, A. B., & Holmes, M. A. (2021). Fiber-type phenotype of the jaw-closing muscles in Gorilla gorilla, Pan troglodytes, and Pan paniscus: A test of the Frequent Recruitment Hypothesis. Journal of Human Evolution, 151, 102938. https://doi.org/10.1016/j.jhevol.2020.102938
Toniolo, L., Cancellara, P., Maccatrozzo, L., Patruno, M., Mascarello, F., & Reggiani, C. (2008). Masticatory myosin unveiled: First determination of contractile parameters of muscle fibers from carnivore jaw muscles. American Journal of Physiology-Cell Physiology, 295(6), C1535–C1542. https://doi.org/10.1152/ajpcell.00093.2008
John Hawks Newsletter
Join the newsletter to receive the latest updates in your inbox.



