Explaining the X chromosome hole in Neandertal ancestry
Natural selection reduced the variation on human X chromosomes in populations with the most Neandertal and Denisovan mixture. It may have been meiotic drive.
A new paper by Laurits Skov and collaborators in Cell Genomics provides a new take on the evolution of recent humans and their relationships with Denisovans and Neandertals. The ancestors of recent people who dispersed from Africa into Eurasia around 70,000 years ago almost immediately experienced widespread strong selective sweeps on the X chromosome.
These repeated selective sweeps help to explain several aspects of human genetic variation on X chromosomes. Over 20 years researchers have tried to explain some of these details of X chromosome diversity in isolation. The new insight from Skov and coworkers suggests a way to make sense of these different observations together, with a shared explanation. The causes of the positive selection remain uncertain, but Skov and coworkers suggest that sex chromosome meiotic drive may be one possibility.
For those of us working to understand human evolution, the interpretation of Neandertal and Denisovan ancestry has to take this new information into account.
X chromosomes have less archaic ancestry
The X chromosomes of recent people have a deficit of Denisovan and Neandertal ancestry compared to the autosomes. I've thought a lot about this observation ever since sequence data started to flow from the bones of these ancient groups. This is the strongest argument for the idea that Neandertals and Denisovans may have been different biological species from today's people.
Nonetheless, I think there are other explanations, and I favor the view that Neandertals and Denisovans were ancient populations of Homo sapiens.
The X chromosome situation is very different from the mitochondrial DNA and Y chromosome. Nobody today has either a Neandertal or Denisovan mtDNA sequence or Y chromosome sequence. Both of these segments of DNA are inherited from one parent only: the mtDNA by all individuals from the mother and the Y chromosome by men from their fathers. That makes both these portions of our DNA outliers compared to the rest of the genome in many ways. Both the mtDNA and the Y chromosome have very recent times to their last common genetic ancestor. For the mtDNA that age is around 250,000 years ago, for the Y chromosome just a bit older at around 300,000 years ago.
Such recent times to their last common ancestor are not very compatible with the diversity of most other parts of the genome. It seems likely that both must have replaced wholescale earlier diversity within ancestral African groups. That replacement was just the latest of earlier episodes, including the wholescale replacement of ancestral Neandertal Y chromosomes and mtDNA by African-derived branches before 250,000 years ago. The most likely cause of such replacements is positive selection, as I pointed out back in 2006 when we had only Neandertal mtDNA to go on.
The X chromosome pattern is more complicated. Many people today have patches of their X chromosomes that descend directly from Denisovan of Neandertal people. But for these living people, the representation of archaic ancestry on the X is only around a fifth of the fraction of Neandertal or Denisovan ancestry found across the autosomes. Additionally, there are some long “introgression deserts”, or regions on the X chromosome where nobody in large samples of living people seem to have Neandertal or Denisovan ancestry.
Laurits Skov and collaborators in a 2020 paper provided the largest-scale picture of introgression in a population today by looking at the genomes of more than 27,000 people from Iceland. In that work, they used a test to find probable introgressed portions of chromosomes similar to either of two Neandertal genomes (Altai and Vindija) and the Denisova 3 genome. At a large scale, the genomes of today's people look like a Jackson Pollock scatter of introgressed DNA from these ancient people. But the X chromosome is different. Introgressed portions from archaic people are much less common, and the ones that occur are found in fewer people.
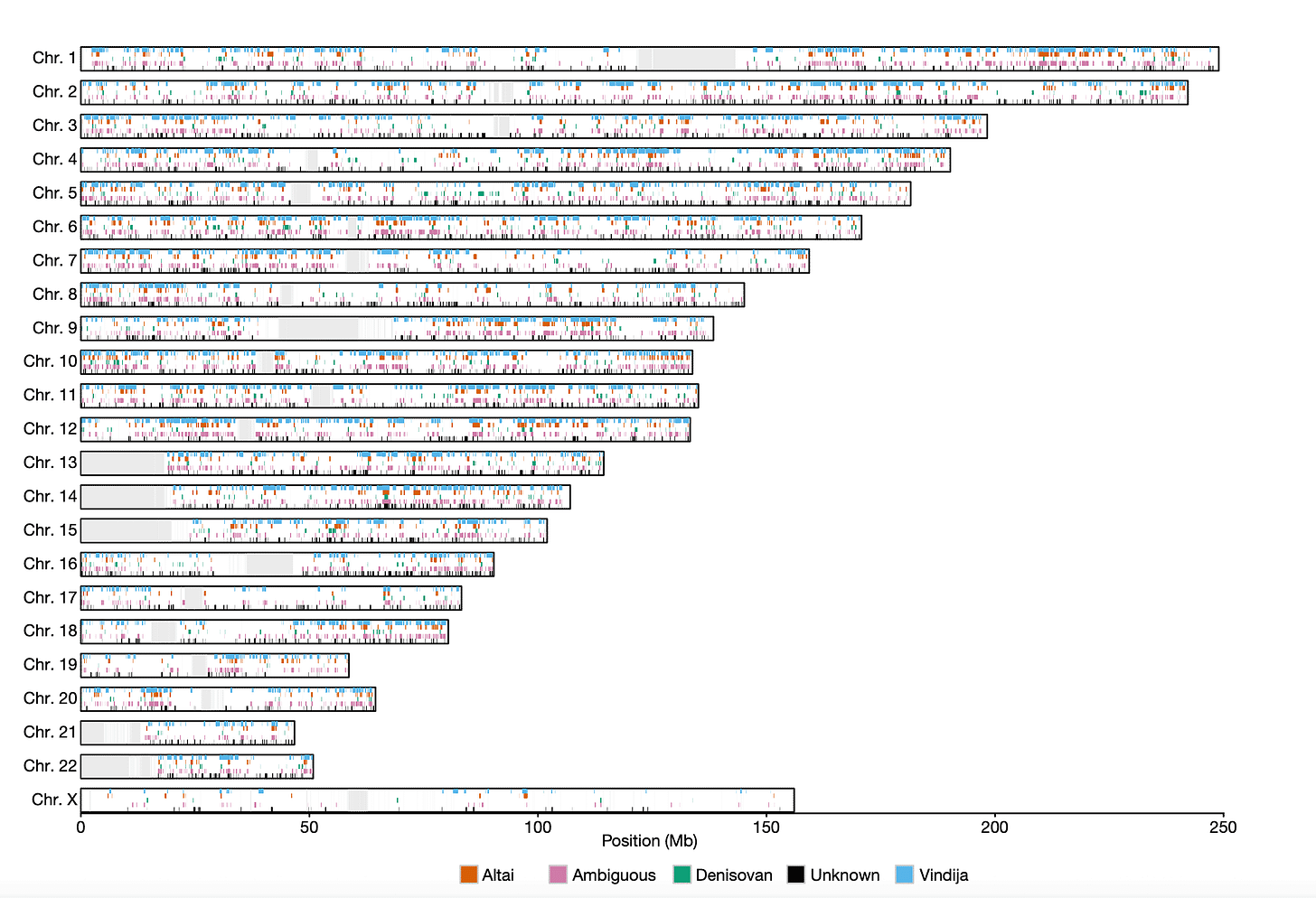
As someone who cares about Denisovans and Neandertals, I've been very interested to understand this pattern. Sriram Sankararaman and coworkers in 2014 were the first to put an accurate number on the lower level of X chromosome introgression, and they pointed to the fact that X chromosome incompatibilities are often seen in hybrids between different species.
“These results suggest that part of the explanation for genomic regions of reduced Neanderthal ancestry is Neanderthal alleles that caused decreased fertility in males when moved to a modern human genetic background.”—Sriram Sankararaman and coworkers
To support this interpretation, Sankararaman and collaborators looked at genes expressed in male testes, which are highly represented on the X chromosome. They found that these genes are strongly underrepresented in Neandertal ancestry. Sankararaman and coworkers suggested that there may have been a connection to male fertility that came into play when Neandertals and the African ancestors of today's people last met and mixed.
I put a high value on this kind of result. If there had been some reproductive challenge for individuals who were hybrids of Neandertal and African ancestry parents, this would be evidence that evolution had erected a partial postzygotic barrier between these groups. Postzygotic barriers are important to our understanding of biological species. When the hybrid offspring of two populations have a pattern of reduced fitness, biologists generally recognize the populations as different biological species.
I have long maintained that paleoanthropologists should rely on the biological species concept when DNA data make it possible. DNA evidence proved that today's humans have Neandertal ancestors in 2010, and for me this evidence of ancient gene flow weighed in favor of recogizing the Neandertals within our own species, Homo sapiens.
But the X chromosome finding dialed back my confidence. If there really was a fitness reduction among hybrids of Neandertals and contemporary African ancestry groups, then the mechanism of speciation must have begun. If this were true, Neandertals would be Homo neanderthalensis after all.
Still, the X chromosome data have not been quite enough to persuade me. For one thing, geneticists kept finding more and more evidence that intermixture among these ancient human groups was neither rare nor unusual. Deeper gene flow from Africa spread Y chromosomes and mtDNA into Neandertals before 100,000 years ago, replacing up to 10% of Neandertal autosomal genome content along the way. The African ancestry groups that dispersed into Eurasia and Oceania encountered and mixed with several genetically diverse Denisovan populations, incorporating new ancestry from each of them. So-called “ghost populations” keep creeping into analyses of today's populations, some even more different from us than the Neandertals.
If X chromosome incompatibilities selected strongly against hybrids, why do we keep finding evidence for hybridization everywhere we look? This doesn't add up.
X chromosomes have less variation outside Africa
A deficit of Neandertal and Denisovan introgression isn't the only interesting thing about X chromosome variation in today's humans. The human X chromosome has a different pattern of variation in populations of Eurasia, Oceania, and the Americas than in sub-Saharan African populations.
For example, the Simons Genome Diversity Project in 2016 provided this figure comparing the X and autosomal heterozygosity in various samples around the world:
Heterozygosity is a measure of genetic variation; it is the probability that an individual is a heterozygote averaged across every site in the genome. In the most diverse human populations in sub-Saharan Africa, this probability is just less than 0.001, meaning that an individual tends to have a single nucleotide polymorphism (SNP) at one out of every 1000 base pairs. In populations in other parts of the world the heterozygosity is lower. It ranges from around 3/4 the sub-Saharan African average in western Eurasia and South Asia, to as low as half that in Indigenous groups of South America.
This is the well-known “diversity cline” of genetic variation across human populations. Africa has the most genetically diverse populations and other populations have less and less, correlated with their distance from Africa. The cline is an echo of the dispersal of African peoples into Eurasia and the rest of the world within the last 100,000 years.
The X chromosome shows an even more extreme cline. Its variation is not only lower within populations far from Africa, it is disproportionately lower.
Some of the lower variation of the X chromosome is explained by its mode of inheritance. Individuals with a Y chromosome have only one copy of the X while they have two copies of all their autosomes, so across the population as a whole there are three copies of the X for every four copies of chromosomes 1 through 22. Based on karyotypes alone, the X should have around 3/4 the variation of the autosomes. When we look at whole genome data, populations in sub-Saharan Africa have X chromosome variation just below that expectation.
But populations in other parts of the world have systematically much lower variation across their X chromosomes, as low as 60–40% of the autosomal variation. Something must have massively reduced X chromosome variation in the ancestors of these groups.
Michael Hammer and coworkers showed in 2010 that the ratio of X chromosome variation to autosomal variation covaries directly with distance to coding regions. Parts of the X chromosome near genes have lower diversity, parts far from genes have diversity more similar to the autosomes. Such a correlation of diversity and functional genome content means that natural selection must be part of the explanation.
In other words, not only Neandertal and Denisovan ancestry but also ancestral African variation was diminished in X chromosomes of peoples outside Africa today. That means that Neandertal and Denisovan mixture were not the causes of this selection, but they may have been casualties of it.
X chromosomes were frequent targets of selection
It is not a new suggestion that positive selection on X chromosome genes might be responsible for lower X chromosome variation in populations outside Africa. Many researchers—including me—were working to understand recent positive selection during the early 2000s.
These studies followed two complementary approaches. One approach examined microarray data of SNP genotypes to identify long linked regions of DNA, known as extended haplotypes, that are at high frequencies in populations today. This pattern can arise when a new mutation comes under strong positive selection and increases rapidly in frequency, bringing a long surrounding region along for the ride.
Pardis Sabeti and coworkers pioneered the extended haplotype approach in 2002, showing that they could successfully demonstrate positive selection on G6PD associated with malaria resistance. Later other teams made more detailed maps of extended haplotypes across the genome, including my collaborators Eric Wang, Robert Moyzis and colleagues in 2006. The next year, my team extended that approach to the International HapMap samples, which led us to the hypothesis that the last 40,000 years of human evolution had seen an acceleration of positive selection on new mutations. At the same time, Benjamin Voight and collaborators were using a similar approach to document extended haplotypes in the Human Genome Diversity Project samples, building a database that could be searched for many years afterward, but now seems to be offline.
These papers found abundant evidence for recent selection on X chromosomes. My team's work had identified genes expressed in testes as a target of selection, a finding that anticipated Sankararaman and coworkers' results on Neandertal introgression.
These early analyses based on microarray data did not make it obvious just how strange the X chromosome is compared to other chromosomes. The institutions that built microarray panels in the early 2000s tried to space polymorphisms evenly across the entire genome. This approach introduced an ascertainment bias that tended to level out low-variation and high-variation regions. What mattered in identifying extended haplotypes was recombination rate, so the X chromosome—which recombines only in mothers—had to be processed differently anyway. As a result, it was hard for us to know at the time whether the X chromosome and autosomes had the same pattern of selection. What mattered from my perspective was that some positive selection was found on all the chromosomes.
A separate line of research from the early 2000s showed pretty strongly that the X chromosome was different from the autosomes when it came to selection. The first high-quality draft of the chimpanzee genome became available in 2005. Rasmus Nielsen and coworkers compared this chimpanzee genome with humans to show that the evolution of human X chromosomes had higher levels of both positive selection and purifying selection than the autosomes during our seven million years of evolution.
Shortly after, Nick Patterson and collaborators showed that the human and chimpanzee X chromosomes had a different pattern of variation at the time of speciation than our autosomes. It seemed that positive selection had swept ancestral hominin and chimpanzee X chromosomes long after their autosomes began to separate from each other. With data from gorillas, it became clear that the reduced difference between human and chimpanzee X chromosomes was a result of many episodes of positive selection in the human-chimpanzee common ancestor. Julien Dutheil and colleagues showed nine wide regions where incomplete lineage sorting is nearly absent from the human-chimpanzee ancestral population, and these align roughly with regions where Neandertal introgression is lowest.
Recurrent positive selection on the X chromosome continued throughout much of hominin evolution. In 2014, Krishna Veeramah and coworkers were able to show that the human X chromosome had been affected by positive selection between 2 and 10 times as often as the autosomes during the span of time since the human-chimpanzee common ancestor.
Selective sweeps began during the dispersal from Africa
The new research from Skov and collaborators focuses on nineteen regions of the X chromosome where at least a fourth of individuals in their sample carry a shared haplotype of 500,000 base pairs or longer. Skov and coworkers suggest that this haplotype length results from positive selection within the last 60,000 years or so. Altogether these regions cover around 11% of the X chromosome.
Extended haplotypes across much of the human genome are quite recent, often dating to the last 10,000 years. These X chromosome extended haplotypes average much older. Skov and coworkers pursued evidence to show that selection dates to a time very soon after the dispersal from Africa. The extended haplotypes are widely shared among all Eurasian, Oceanian, and American samples, and nine of them are present in the genome of the 45,000-year-old Ust'-Ishim individual, meaning at that time the extended haplotypes must already have been at frequencies similar to today.
It's striking how ubiquitous these extended haplotypes are across Eurasian, Oceanian, and American populations. Within the smaller sample of African populations examined by Skov and coworkers, these extended haplotypes are entirely absent.

What this means is that the positive selection on these haplotypes must have been underway by the time the dispersing African populations met Neandertals. These haplotypes were already at or near their current frequencies when these populations mixed with Denisovans.
That raises the question: Was this positive selection entirely responsible for the reduced fraction of Neandertal and Denisovan ancestry on the X chromosome? Skov and coworkers show that this does indeed appear to be the case—at least for the regions with the extended haplotypes. The extended haplotypes have almost no evidence of Neandertal or Denisovan ancestry. By contrast, copies of the X in the same population samples that lack the extended haplotypes have just as much archaic ancestry as the X chromosome as a whole.

For these regions at least, it was not purifying selection against hybrids that reduced archaic introgression. It was the positive selection in favor of a few extended haplotypes in different parts of the X chromosome.
That's a very satisfying result. Still, it doesn't explain the entire pattern of reduced Neandertal and Denisovan ancestry across the X chromosome. There are still some introgression deserts with very low archaic ancestry, that may have nothing to do with the extended haplotypes. But widespread selection on multiple haplotypes in the early ancestors of non-African populations could easily have effects beyond the tightly linked extended haplotypes themselves. I suspect that the pattern has nothing to do with archaic people and everything to do with the unique circumstances of population dispersal.
Meiotic drive on sex chromosomes?
So what adaptive purpose did these extended haplotypes serve? Skov and collaborators considered this question but did not come to any strong answer. There is no category of functional genes that seems enriched in the regions covered by the extended haplotypes. It is possible that there were many different causes of selection on individual genes that all just happened to make a difference in this founder population.
On the other hand, this is a lot of selection in a population that started small.
One clue comes from comparing the distribution of the extended haplotypes with earlier evidence from comparative genomics. Julien Dutheil and coworkers in 2013 studied the pattern of incomplete lineage sorting among human, chimpanzee, and gorilla genomes.

Considering what they could compare, Skov and coworkers suggested that the X chromosome pattern of positive selection may have something to do with the functional genetics of sex determination.
“At the present time, we cannot envision a likely scenario that explains all our observations. We cautiously hypothesize that the selective sweeps may be due to sex-chromosome meiotic drive: if an averagely even transmission of X and Y sperm in meiosis is maintained by a dynamic equilibrium of antagonizing drivers on X and Y, the bottlenecked main out-of-Africa population may have been invaded by sex-chromosome drivers retained in an earlier out-of-Africa population.”—Laurits Skov and coworkers
From this point of view, both X and Y chromosomes from fathers find themselves in a race to be represented in the population of sperm cells. Any gene on the X that can prevent the formation of Y-bearing sperm will tend to increase in frequency, and vice-versa. This phenomenon is known as meiotic drive.
When it happens on the sex chromosomes, meiotic drive has the side-effect of increasing the ratio of the sex determined by the driving chromosome. A driving X chromosome gene will increase the fraction of girls born in the population. Eventually, a population where many men have more daughters will confer a reproductive advantage on fathers who can produce Y chromosome-bearing sperm, and vice-versa. Hence, sex-linked meiotic drive is limited in the long run by antagonistic selection.
The scenario is an appealing one for several reasons. First, it helps to explain why the X chromosome extended haplotypes seem to have stalled out without becoming fixed everywhere outside Africa—or for that matter, within Africa. There must be some kind of antagonistic selection or diminishing returns at play to explain why these have not swept to fixation. Second, the scenario makes sense of the historically rapid fixation of substitutions on the X chromosome and the reduction of incomplete lineage sorting across the X chromosomes of human-chimpanzee and other great ape ancestors.
The third reason may be more provocative. Human evolution during the last 50,000 years has included the rapid increase in frequency of many Y chromosome haplogroups. Some of these have spread across massive land areas, like the R1a and R1b lineages across much of western and central Eurasia. Geneticists have mostly assumed that such haplogroups were the beneficiaries of founder effects, as initially small populations expanded together with languages, subsistence innovations, or conquering hordes. But Y chromosomes have genes that may also be subject to positive selection. Maybe some Y chromosome haplogroups have arisen as responses to X chromosome genes that have been tinkering with testes.
Still, there's one big reason to hesitate before accepting a meiotic drive explanation. Humans today show little or no sign of additive genetic variance in the sex ratio born either to fathers or mothers. The best current data come from Sweden, where a dataset including millions of births to siblings shows no significant heritability of sex ratio. If there were segregating variants on the X chromosome that became common by driving changes to X and Y chromosome representation in sperm (or egg) populations, they would surely show up as heritable causes of variation in the sex ratio of children at birth.
To me that isn't a fully definitive rejection of the idea of meiotic drive. The X chromosome variants did, after all, evidently stall out, suggesting that whatever effects they had in early humans have been attenuated in recent populations. Few people in Sweden lack these extended haplotypes. Maybe the landscape of antagonistic selection is complex enough that such variants largely cancel out today.
Bottom line
The hypothesis will be testable in the long run, and I'll be interested to see how it plays out.
In the meantime, the results of the Skov study matter a lot to our interpretation of Neandertal and Denisovan ancestry. The X chromosome was not a favorable ground for the genes of these ancient people. The early expansion of African people into Eurasia was accompanied by positive selection on many X chromosome extended haplotypes, different from the pattern in African populations. That evolutionary trajectory excluded Neandertal and Denisovan ancestry, and at the same time eliminated a large fraction of African ancestry from the X chromosome.
I return to the question of Neandertals and Denisovans as possible species. It may seem like only a semantic difference whether ancient gene flow occurred among populations of a single species or by means of hybridization between species. Whether today's anthropologists can use the name Homo neanderthalensis for Neandertals cannot possibly matter to these ancient people. They are long gone and whatever ideas they had about themselves are beyond recovering. Nor does it change anything about their legacy, which lives within today's people whatever we call them.
But the genetic mechanisms of our origins matter. They matter to how we understand our evolutionary potential, and the potential of our ancestors. Was the potential of ancient people circumscribed by their evolutionary history, or could they join other populations without limits?
I don't think we know the answer to this yet.
Notes: This post digs deep into my history in understanding selection in human populations. I've cited below many of the key articles but obviously this is a huge area of research from the early 2000s and 2010s that has progressed greatly since then. There's much more to write along these lines.
The work on the heritability of sex ratio in offspring is from Zietsch and coworkers (2020).
References
Dutheil, J. Y., Munch, K., Nam, K., Mailund, T., & Schierup, M. H. (2015). Strong Selective Sweeps on the X Chromosome in the Human-Chimpanzee Ancestor Explain Its Low Divergence. PLOS Genetics, 11(8), e1005451. https://doi.org/10.1371/journal.pgen.1005451
Hammer, M. F., Woerner, A. E., Mendez, F. L., Watkins, J. C., Cox, M. P., & Wall, J. D. (2010). The ratio of human X chromosome to autosome diversity is positively correlated with genetic distance from genes. Nature Genetics, 42(10), Article 10. https://doi.org/10.1038/ng.651
Hawks, J. (2006). Selection on mitochondrial DNA and the Neanderthal problem. In J.-J. Hublin, K. Harvati, & T. Harrison (Eds.), Neanderthals Revisited: New Approaches and Perspectives (pp. 221–238). Springer Netherlands. https://doi.org/10.1007/978-1-4020-5121-0_12
Hawks, J., Wang, E. T., Cochran, G. M., Harpending, H. C., & Moyzis, R. K. (2007). Recent acceleration of human adaptive evolution. Proceedings of the National Academy of Sciences, 104(52), 20753–20758. https://doi.org/10.1073/pnas.0707650104
Nielsen, R., Bustamante, C., Clark, A. G., Glanowski, S., Sackton, T. B., Hubisz, M. J., Fledel-Alon, A., Tanenbaum, D. M., Civello, D., White, T. J., Sninsky, J. J., Adams, M. D., & Cargill, M. (2005). A Scan for Positively Selected Genes in the Genomes of Humans and Chimpanzees. PLOS Biology, 3(6), e170. https://doi.org/10.1371/journal.pbio.0030170
Patterson, N., Richter, D. J., Gnerre, S., Lander, E. S., & Reich, D. (2006). Genetic evidence for complex speciation of humans and chimpanzees. Nature, 441(7097), Article 7097. https://doi.org/10.1038/nature04789
Sabeti, P. C., Reich, D. E., Higgins, J. M., Levine, H. Z. P., Richter, D. J., Schaffner, S. F., Gabriel, S. B., Platko, J. V., Patterson, N. J., McDonald, G. J., Ackerman, H. C., Campbell, S. J., Altshuler, D., Cooper, R., Kwiatkowski, D., Ward, R., & Lander, E. S. (2002). Detecting recent positive selection in the human genome from haplotype structure. Nature, 419(6909), Article 6909. https://doi.org/10.1038/nature01140
Sankararaman, S., Mallick, S., Dannemann, M., Prüfer, K., Kelso, J., Pääbo, S., Patterson, N., & Reich, D. (2014). The genomic landscape of Neanderthal ancestry in present-day humans. Nature, 507(7492), Article 7492. https://doi.org/10.1038/nature12961
Skov, L., Macià, M. C., Lucotte, E. A., Cavassim, M. I. A., Castellano, D., Schierup, M. H., & Munch, K. (2023). Extraordinary selection on the human X chromosome associated with archaic admixture. Cell Genomics, in press. https://doi.org/10.1016/j.xgen.2023.100274
Skov, L., Coll Macià, M., Sveinbjörnsson, G., Mafessoni, F., Lucotte, E. A., Einarsdóttir, M. S., Jonsson, H., Halldorsson, B., Gudbjartsson, D. F., Helgason, A., Schierup, M. H., & Stefansson, K. (2020). The nature of Neanderthal introgression revealed by 27,566 Icelandic genomes. Nature, 582(7810), Article 7810. https://doi.org/10.1038/s41586-020-2225-9
Veeramah, K. R., Gutenkunst, R. N., Woerner, A. E., Watkins, J. C., & Hammer, M. F. (2014). Evidence for Increased Levels of Positive and Negative Selection on the X Chromosome versus Autosomes in Humans. Molecular Biology and Evolution, 31(9), 2267–2282. https://doi.org/10.1093/molbev/msu166
Wang, E. T., Kodama, G., Baldi, P., & Moyzis, R. K. (2006). Global landscape of recent inferred Darwinian selection for Homo sapiens. Proceedings of the National Academy of Sciences, 103(1), 135–140. https://doi.org/10.1073/pnas.0509691102
Zietsch, B. P., Walum, H., Lichtenstein, P., Verweij, K. J. H., & Kuja-Halkola, R. (2020). No genetic contribution to variation in human offspring sex ratio: A total population study of 4.7 million births. Proceedings of the Royal Society B: Biological Sciences, 287(1921), 20192849. https://doi.org/10.1098/rspb.2019.2849